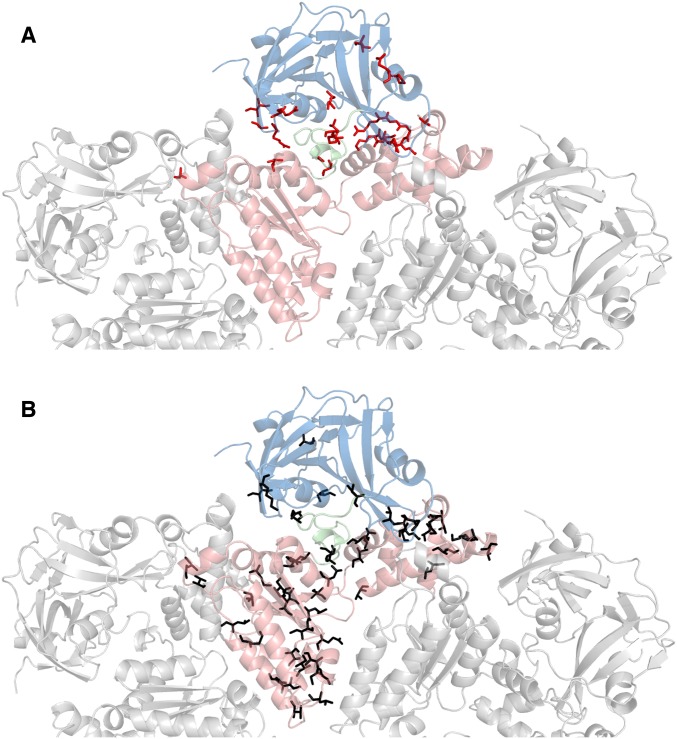
Fig. S2.
Location of disease-associated mutations (A) (12) and predominantly hydrophobic substitutions that have been tested (B) in ND1Lp97. Top view of a half-hexamer of ND1Lp97-ADP in cartoon representation with the central protomer color-coded by domain (NTD-linker-D1 in blue, green, red). (A) The 22 residues in ND1Lp97 that are mutated in disease patients (12) are shown in red (sticks) on the structure of the ADP state [PDB ID code 1E32 (33)]. (B) Black sticks denote the positions of 56 point mutations that do not shift the NTD up/down equilibrium in p97-ADP.