Figure 4.
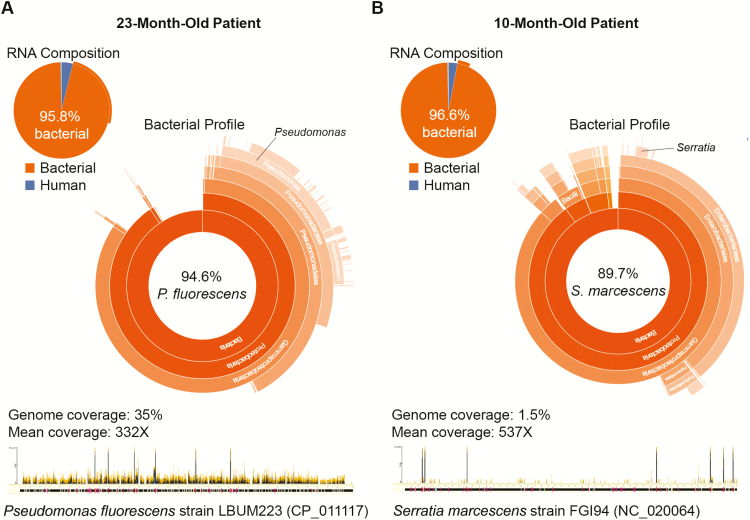
An abundant bacterial flora (>95% of sequencing reads) dominated by a single potential pathogen was detected by RNA sequencing in nasopharyngeal/oropharyngeal (NP/OP) samples of 2 children with community-acquired pneumonia and no identified pathogen by the Etiology of Pneumonia in the Community study protocol. A, In a 23-month-old patient, 94.6% of sequencing reads generated from the NP/OP sample was identified as Pseudomonas fluorescens, covering 35% of the genome of strain LBUM223 (NCBI accession number CP_011117) at a mean of 332X (data analyzed as described in [14]). B, In a 10-month-old patient, 89.7% of sequencing reads were derived from Serratia marcescens, covering 1.5% of the genome sequence of strain FGI94 (NCBI accession number NC_020064) at a mean of 537X.