Fig. 3. Universal bacteria detection.
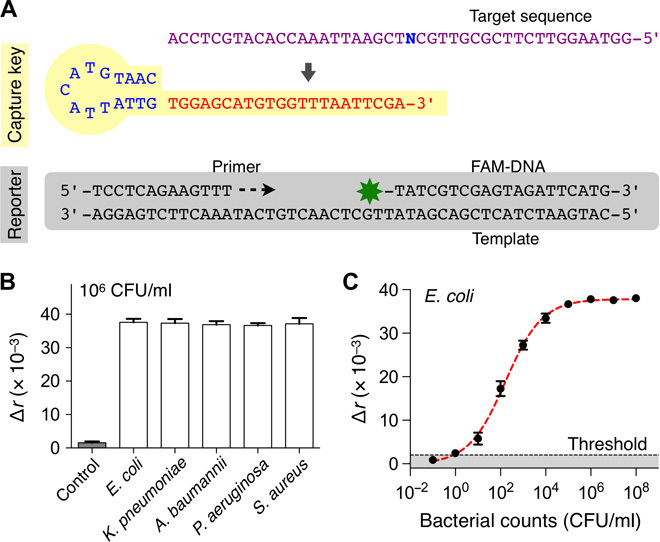
(A) The universal capture key (UNI key) detects a conserved bacterial sequence (N = A for Escherichia, Klebsiella, and Acinetobacter; N = T for Pseudomonas and Staphylococcus). The reporter is composed of a primer, a template, and FAM-DNA. (B) Five different HAI pathogens (106 CFU/ml) were detected with the PAD. The signal levels were statistically identical among concentration-matched bacterial samples. (C) Detection sensitivity. Samples with different bacterial concentrations (E. coli, 10–1 to 108 CFU/ml) were prepared through serial dilution. The limit of detection (LOD) was at the level of a single bacterium. The threshold was set at 3× SD above background signal of the sample without bacteria. All experiments were performed in triplicate and the graphs are displayed as means ± SD.
