Chronic lymphocytic leukemia (CLL), the most prevalent form of leukemia in adults in the western world, has a highly heterogeneous clinical course, and is characterized by genomic instability which gives rise to several chromosomal alterations detectable in more than 80% of CLL cases.1 While the absence of somatic mutations in the immunoglobulin heavy-chain gene variable region (IGVH) and high expression of CD38 and ZAP-70 molecules have been associated with aggressive CLL, the most common chromosomal abnormality, 13q14 deletion (13q14del), has been associated with a more indolent form of the disease.1,2 The mechanism by which 13q14del contributes to CLL pathogenesis and affects the outcome of patients has not yet been elucidated.
Several studies have focused on the prognostic significance of an interplay between telomeres and telomerase in CLL.3–6 Telomerase is responsible for the maintenance of telomeres, structures which cap and protect the ends of chromosomes. Telomerase expression, up-regulated in approximately 90% of human cancers, enables continuous and uncontrolled proliferation of the malignant cells driving tumor growth and progression.7 Catalytic protein with telomere-specific reverse transcriptase activity (TERT) is the rate-limiting component of the telomerase complex, and its expression is correlated with telomerase activity.8 We and others have shown that levels of TERT and its activity are prognostic markers in CLL.3–6 Notably, TERT levels were found to be lower in 13q14del CLL than in CLL with other chromosomal abnormalities,4 but mechanistic insight for this difference is still unclear.
The expression of microRNA, small non-coding RNA with regulatory functions, is frequently deregulated in tumors.9 The microRNA cluster which encodes for miR-15a and miR-16-1 maps within a 30-kilobase region of loss at 13q14.10 Both these microRNA interact directly with and inhibit the expression of the anti-apoptotic BCL2 gene, and the loss of the miR-15a/miR-16-1 cluster due to 13q14 deletion is the main cause of BCL2 overexpression in CLL.10 Nonetheless, miR-15a and miR-16-1 also directly target the tumor suppressor TP53 gene, and their overexpression is associated with repressed TP53 expression at both mRNA and protein levels.11 Transcription of the TERT gene is the key determinant in regulating TERT expression and telomerase activity. TP53 is an important repressor of the promoter of TERT gene,12 and the C-terminus of TP53 interacts with and inhibits telomerase activity.13
This study aimed at determining miR-15a/miR16-1 levels in CLL and correlating them to TP53 and TERT transcripts. Since 13q14del CLL cells would lack miR-15a/miR-16-1, and since these two microRNA repress expression of TP53,11 we hypothesized that the miRNA/TP53 axis modulates TERT levels, and thus the outcome of patients with 13q14del CLL.
Peripheral blood cells were collected from 155 CLL patients who attended the Hematology Section, Clinical and Experimental Medicine, University of Padua (Italy). This study included 99 cases of CLL with the 13q14 deletion as the sole chromosomal abnormality detected by fluorescence in situ hybridization (FISH), and 56 CLL with no chromosomal abnormalities detected by FISH [i.e., 13q14.3, 17p13.1 (TP53), and 11q22.3 (ATM) deletions, and trisomy 12]. FISH was carried out as previously described.4 CLL cases with TP53 mutations were excluded from this study. All samples were collected at the time of diagnosis, and all patients were untreated at the time of sampling. TERT transcripts were quantified in all samples by real-time polymerase chain reaction, as previously described.4 The expression of TP53 transcripts was quantified in 123 CLL by real-time polymerase chain reaction using a TaqMan Gene Expression Assay-Human TP53:Hs01034249_m1 kit (Life Technologies, Carlsbad, CA, USA), following the manufacturer’s instructions. The expression of miR-15a, miR-16-1 and RNU6B, used as a control to normalize the data, was assessed in available RNA samples from 101 cases of CLL using a standard TaqMan MicroRNA assay kit (Life Technologies), according to the manufacturer’s instructions. Statistical analyses were carried out using SAS version 9.1 (SAS Institute, Cary, NC, USA). Time from diagnosis to first treatment was considered as a marker for time to disease progression. Informed consent was obtained according to the Helsinki Declaration and the study was approved by the local Ethics Committee.
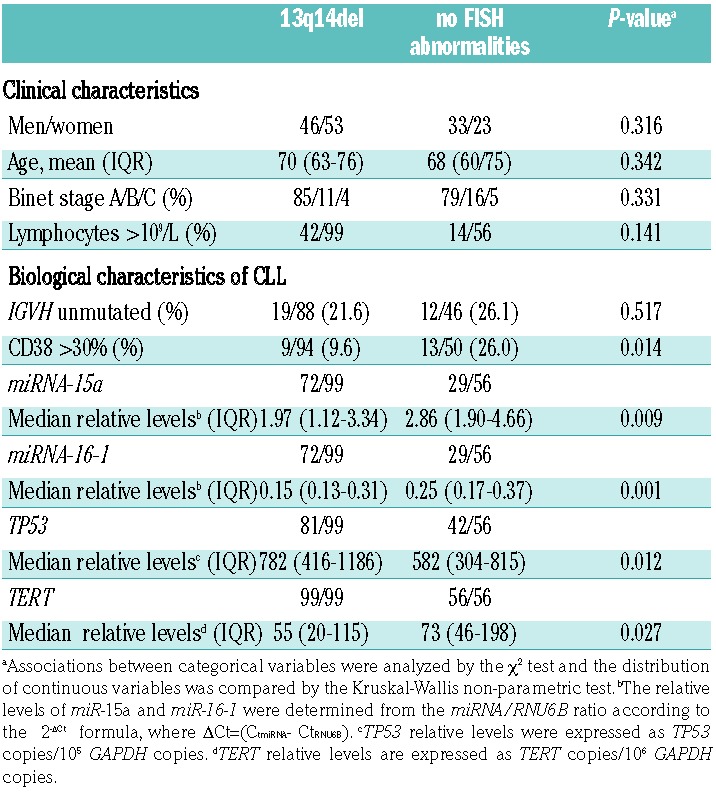
As expected, both miR-15a and miR-16-1 levels were lower in 13q14del CLL than in CLL without chromosomal abnormalities [1.97 (1.12–3.34) versus 2.86 (1.90–4.66), P=0.009 and 0.15 (0.13–0.31) versus 0.25 (0.17–0.37), P=0.001)] (Table 1) and positive correlations were found between miR-15a and miR-16-1 levels (rs=0.854, P<0.0001). In agreement with the fact that overexpression of miR-15a and miR-16-1 in primary CLL cells is associated with a decrease in TP53 levels,11 TP53 mRNA levels were significantly higher in 13q14del CLL than in CLL with no chromosomal abnormalities [782 (416–1186) versus 582 (304–815) copies/105 GAPDH copies P=0.012] (Table 1). Levels of miR-15a and miR-16-1 were inversely correlated with TP53 levels in both 13q14del (rs= −0.775, P<0.0001 and rs= −0.722, P<0.0001, respectively; data not shown) and all CLL (rs= −0.806, P<0.0001 and rs= −704, P<0.0001, respectively; data not shown).
Table 1.
Clinical and biological characteristics of the patients with chronic lymphocytic leukemia according to 13q14del status.
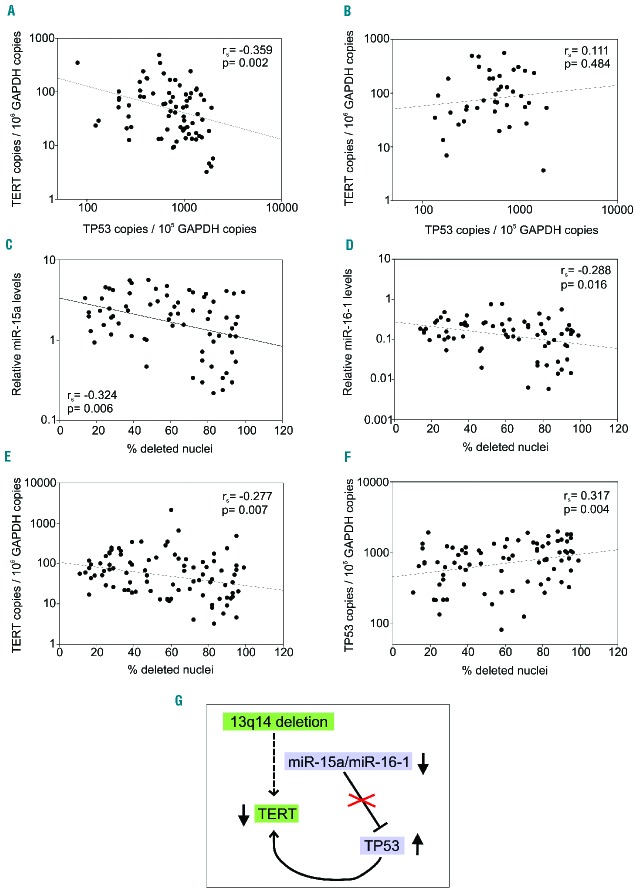
13q14del CLL expressed significantly lower levels of TERT than CLL with normal cytogenetic profile [55 (20–115) versus 73 (46–198) copies/106 copies GAPDH, P=0.027] (Table 1). Of interest, TP53 levels correlated negatively with TERT levels only in 13q14del (rs= −0.359, P=0.002, Figure 1A), but not in CLL without chromosomal abnormalities (rs= 0.111, P=0.484) (Figure 1B). The different percentage of nuclei carrying the 13q14 deletion may explain the variable levels of miRNA, TP53, and TERT observed within the 13q14del CLL; indeed, the percentage of nuclei carrying the 13q14 deletion tended to correlate negatively with miR-15a, miR-16-1 and TERT levels (rs= −0.324, P=0.006; rs= −0.288, P=0.016; rs= −0.277, P=0.007, respectively) (Figure 1C–E), and positively with TP53 levels (rs=0.317, P=0.004) (Figure 1F).
Figure 1.
Relationship among miRNA, TP53 and TERT levels in 13q14del chronic lymphocytic leukemia. (A, B) Relationship between TERT and TP53 levels in: (A) 13q14del CLL, (B) CLL with no chromosomal abnormalities. (C-F) Relationship between percentage of deleted nuclei and levels of (C) miR-15a, (D) miR-16-1, (E) TP53, (F) TERT. (G) Proposed network among miR-15a/miR-16-1, TP53, and TERT expression. Down-regulation of miR-15a/miR-16-1, due to 13q14 deletion, leads to increased TP53 levels which, in turn, down-regulate levels of TERT, the catalytic component of the telomerase complex.
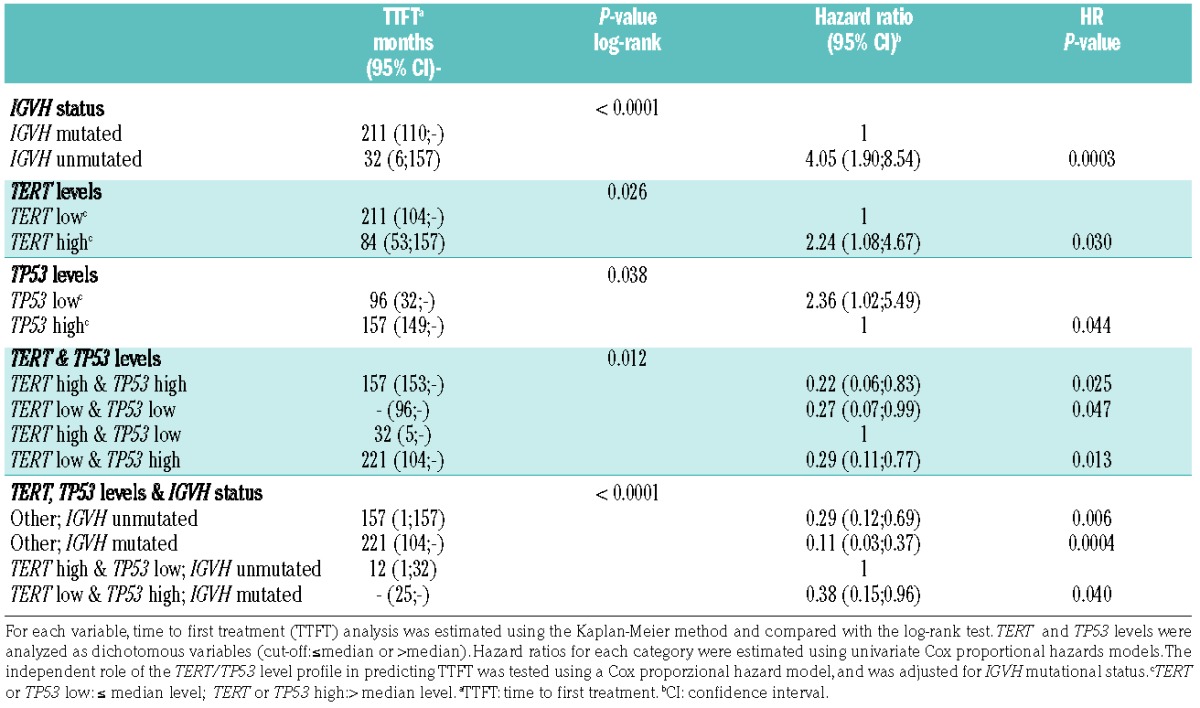
Univariate Cox analyses showed that mutated IGVH status and low TERT levels (≤ median) were prognostic of better disease outcome, estimated as time from diagnosis to first treatment, in the entire cohort of CLL, with hazard ratios (HR) and 95 confidence intervals (95% CI) of 3.95 (2.11–7.43) P<0.001 and 2.19 (1.21–3.97) P=0.008, respectively, and also in the subgroup of patients with 13q14del CLL (Table 2). Of interest, cases with high TP53 expression had a better prognosis in the subgroup of 13q14del CLL (Table 2), but not in that of CLL with normal cytogenetic profile [HR 0.97 (95% CI 0.29–3.28); P=0.973]. CD38 (>30%) was not prognostic of disease outcome in 13q14del CLL [HR 2.28 (95% CI 0.86–6.04) P=0.107; data not shown]. Multivariate Cox analysis showed that within 13q14del CLL, a high TERT/low TP53 level profile defined the subgroup of cases with the worst prognosis (Table 2) and the values of TERT/TP53 profiles were independent of IGVH mutational status (Table 2).
Table 2.
Time to first treatment and hazard ratio according to IGVH status, TERT levels, and TP53 levels showing their effects on disease progression in chronic lymphocytic leukemia with 13q14 deletion.
This result, together with the finding that a negative correlation between TP53 and TERT levels can only be observed in 13q14del CLL but not in CLL without chromosomal abnormalities, emphasizes the existence of a network between miR-15a/miR-16-1, TP53 and TERT within 13q14del CLL (Figure 1G).
It should be noted that miRNA expression also varied in CLL without chromosomal aberrations; therefore, mechanisms other than deletion of the miRNA cluster in the 13q14 region probably contribute to regulating their expression. It has recently been demonstrated that miR-15a/miR16-1 and TP53 are engaged in a feedback loop in CLL: increased levels of miR-15a/miR16-1 target and down-regulate TP53 expression, while TP53 binds to its specific binding sites on chromosome 13 and up-regulates the expression of miR-15a/miR-16-1 in CLL with a normal cytogenetic profile.11 As the expression of TP53 in CLL is influenced by many factors,14 this variability may influence levels of the miRNA, which in turn down-regulate TP53 expression.
Notably, in this study TP53 had prognostic value only for 13q14del CLL. It has been advanced that CLL with a high percentage of 13q14 deletion tend to have a worse prognosis.15 The deletion of miR-15a and miR-16-1 and the consequent lack of inhibition of the anti-apoptotic BCL2 gene may partially support this trend. However, miR-15a and miR-16-1 also target the tumor suppressor gene TP53. Hence, the loss of the miR-15a/miR-16-1 cluster due to 13q14 deletion not only moves the balance toward higher levels of anti-apoptotic protein, but also toward higher levels of tumor suppressor TP53. With a cut-off of 80% of 13q14 deleted nuclei,15 we did not find any significant differences in disease outcome (HR 0.597; 95% CI: 0.304–1.173; P=0.135). This supports the concept that the effects of deletion, rather than the percentage of deletion per se, influence the disease outcome.
An interesting and intriguing result of our study is that levels of TP53 were inversely correlated with TERT levels only in the 13q14del CLL cases. TP53 alone may be inefficient in regulating TERT, since many other factors activate or repress TERT at a transcriptional level.7 Our findings suggest that TP53 plays an important role in TERT regulation in 13q14del CLL, while in CLL with no or other chromosomal aberrations TERT may be regulated by other factors.
In conclusion, collectively, these findings indicate that, in 13q14del CLL, the miR15/miR16-1 cluster and TP53 axis is an important pathway which regulates TERT expression, thus influencing disease outcome, and also suggest that analysis of TERT/TP53 profiles may be useful in refining the prognosis of patients with 13q14del CLL.
Supplementary Material
Acknowledgments
The authors would like to thank all the patients who provided samples for the study.
Footnotes
Funding: this study was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC grant n. 14258) and by Istituto Oncologico Veneto
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. [DOI] [PubMed] [Google Scholar]
- 2.Nabhan C, Raca G, Wang L. Predicting prognosis in chronic lymphocytic leukemia in the contemporary era. JAMA Oncol. 2015;1(7):965–974. [DOI] [PubMed] [Google Scholar]
- 3.Terrin L, Trentin L, Degan M, et al. Telomerase expression in B-cell chronic lymphocytic leukemia predicts survival and delineates subgroups of patients with the same igVH mutation status and different outcome. Leukemia. 2007;21(5):965–972. [DOI] [PubMed] [Google Scholar]
- 4.Rampazzo E, Bonaldi L, Trentin L, et al. Telomere length and telomerase levels delineate subgroups of B-cell chronic lymphocytic leukemia with different biological characteristics and clinical outcomes. Haematologica. 2012;97(1):56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palma M, Parker A, Hojjat-Farsangi M, et al. Telomere length and expression of human telomerase reverse transcriptase splice variants in chronic lymphocytic leukemia. Exp Hematol. 2013;41(7):615–626. [DOI] [PubMed] [Google Scholar]
- 6.Hoxha M, Fabris S, Agnelli L, et al. Relevance of telomere/telomerase system impairment in early stage chronic lymphocytic leukemia. Gene Chromosome Canc. 2014;53(7):612–621. [DOI] [PubMed] [Google Scholar]
- 7.Dolcetti R, De Rossi A. Telomere/telomerase interplay in virus-driven and virus-independent lymphomagenesis: pathogenic and clinical implications. Med Res Rev. 2012;32(2):233–253. [DOI] [PubMed] [Google Scholar]
- 8.Daniel M, Peek GW, Tollefsbol TO. Regulation of the human catalytic subunit of telomerase (hTERT). Gene. 2012; 498(2):135–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008; 14(1):1–6. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002; 99(24):15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabbri M, Bottoni A, Shimizu M, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA 2011;305(1):59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu D, Wang Q, Gruber A, et al. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19(45):5123–5133. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Cao Y, Berndt MC, Funder JW, Liu JP. Molecular interactions between telomerase and the tumor suppressor protein p53 in vitro. Oncogene. 1999;18(48):6785–6794. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, Wang X. The role of TP53 network in the pathogenesis of chronic lymphocytic leukemia. Int J Clin Exp Pathol. 2013;6:1223–1229 [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez AE, Hernandez JA, Benito R, et al. Molecular characterization of chronic lymphocytic leukemia patients with a high number of losses in 13q14. PloS One. 2012;7(11):e48485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.