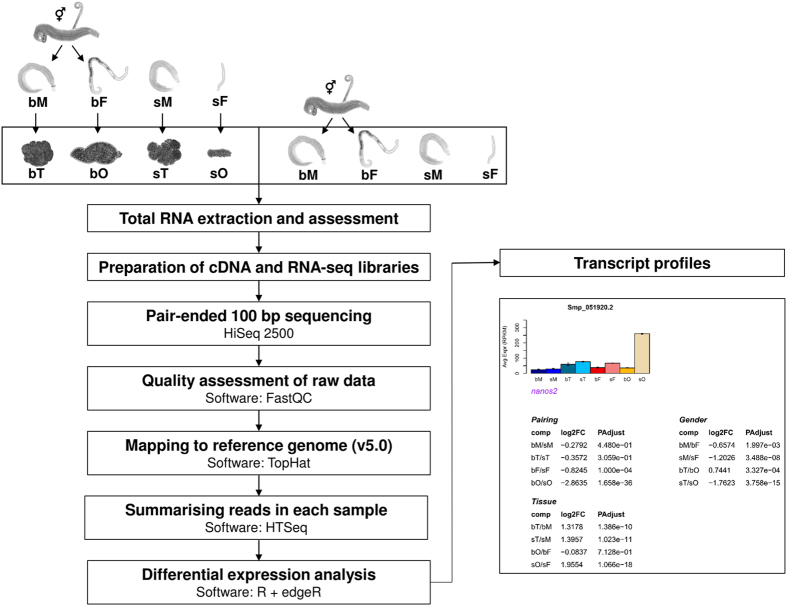
Figure 1. Schematic overview of the experimental design.
All biological replicates (n=2 for sO and n=3 for all other samples) were obtained from the species Schistosoma mansoni, which was maintained in hamsters as final hosts. The separation of couples and the isolation of ovaries and testes were always performed at the same day to avoid any in vitro-culture effects. Total RNA was extracted by Trizol and its quality assessed by electropherogram analyses using an Agilent 2100 Bioanalyzer and Pico chips (Agilent Technologies). For each sample 100 ng of total RNA was used for synthesizing cDNA and generating libraries. Pair-end 100 bp sequencing was performed on Illumina HiSeq 2500 running two technical replicates for each sample. Raw reads were assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and mapped to Schistosoma mansoni reference genome (v5.0) using TopHat. Reads for each transcript were counted using HTSeq and imported to edgeR for normalization across all samples and differential expression analysis. Mean RPKM and s.e.m. values based on normalized reads were calculated and used for barplots. A transcript profile was generated for each gene, including the barplot, as well as the log2 fold-change (log2FC) and adjusted P (PAdjust) values based on various comparisons (see Smp_051920.2 as representative example, a nanos-ortholog that is abundantly transcribed in ovaries of unpaired females=sO).