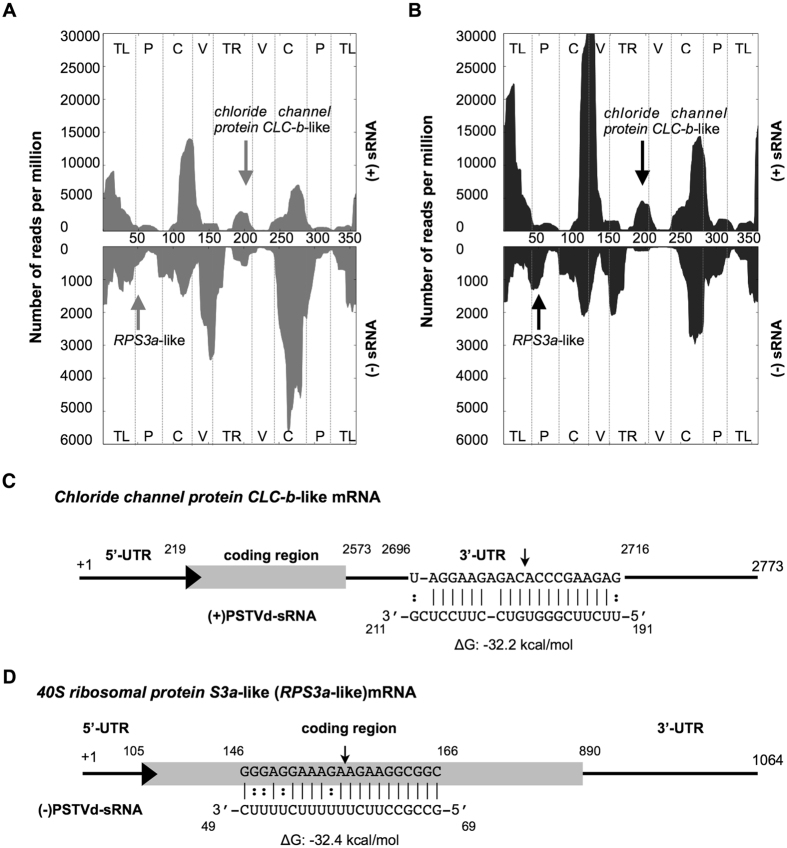
Figure 1.
Profiling of the vd-sRNAs recovered from PSTVd infected tomato plants and the predicted vd-sRNA:target duplexes. Sequence profiles of the (+) and the (−) PSTVd-sRNA populations recovered from the leaf tissues of infected tomato plants. Panels (A) and (B) represent the profiles of the (+) and the (−) PSTVd-I derived sRNAs, while panels (C) and (D) represent the profiles of the (+) and the (−) PSTVd-RG1 derived sRNAs. The vertical arrows denote the two vd-sRNA populations of particular interest, namely those capable of targeting potential host mRNAs: a (+) vd-sRNA located at positions 119 to 211 and a (−) vd-sRNA located at positions 69 to 49. Please note that different scales are used so as to compensate for the lower numbers of (−) vd-sRNA sequences recovered for both of the PSTVd variant infected plants. The predicted interactions between the PSTVd-sRNAs derived from the (C) (+) strand with the 3′ UTR of the chloride channel protein CLC-b-like mRNA and (D) the (−) strand with the coding region of the 40S ribosomal protein S3a-like mRNA are shown. The arrows indicate the predicted RISC mediated cleavage sites. The sequences are shown in the complementary polarity. The PairFold online tool was used to predict the minimum free energy (ΔG) secondary structures of the pairs of RNA sequences.