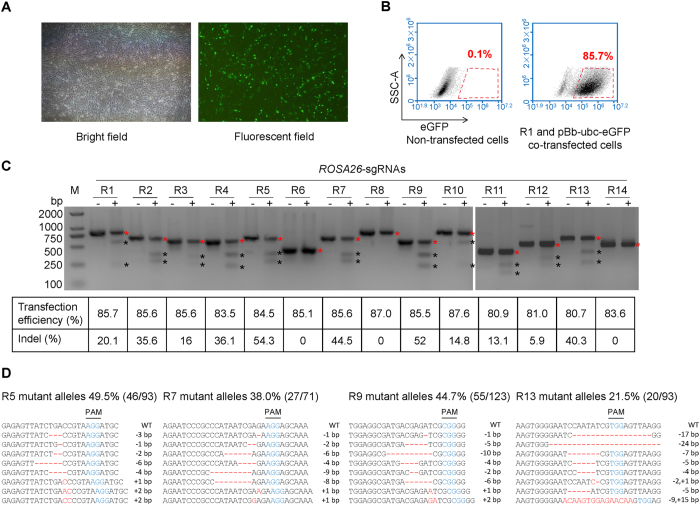
Figure 1.
CRISPR/Cas9 induced targeted mutations at the ROSA26 locus. (A) Fluorescence expression in porcine fetal fibroblasts co-transfected with sgRNAs and pBb-ubc-eGFP by electroporation. (B) Detection of transfection efficiency by flow cytometry at 48 h post-transfection. (C) T7E1 assay showing percentage of indel formations (+) in the 14 targeted sites of pig ROSA26 locus. Control cells (−) were transfected with Cas9 only (without sgRNA). The transfection efficiency and indel percentages for each sgRNA are shown at the bottom. Representative images from n = 3 independent experiments. The red and black asterisks indicate uncut and cut bands by T7E1, respectively. (D) Sanger sequencing detected targeted mutation (indels) at R5, R7, R9, and R13 targeted sites in ROSA26. Eight mutant alleles are presented as representative of all the mutant alleles for each sgRNA transfected group. The fractions in the brackets indicate the ratios of mutant alleles to all sequenced alleles. Indels are shown in red letters and dashes. PAM sequence is shown in blue.