Figure 6.
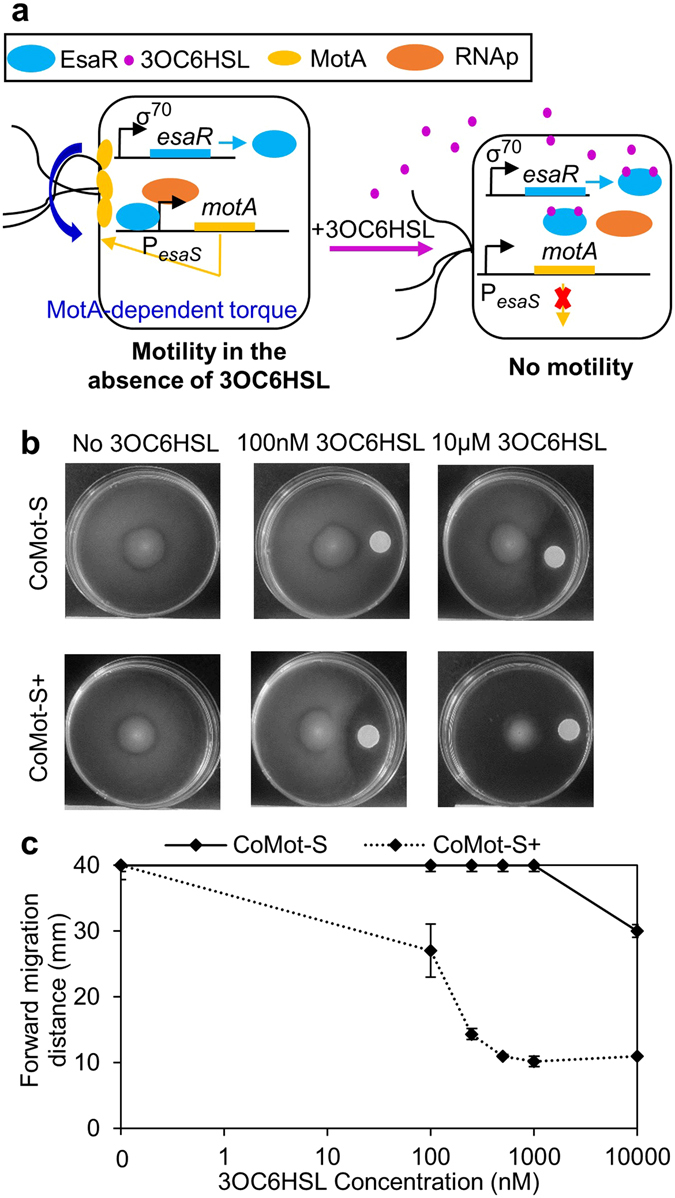
Characterization of the migration response of CoMot-S and CoMot-S+ cells in a 3OC6HSL gradient shows directional movement of cells away from a 3OC6HSL source: (a) Illustration of 3OC6HSL-dependent motA expression in the CoMot-S strain. Expression of motA is under the control of the PesaS promoter. esaR is constitutively expressed from a σ70-dependent promoter and activates expression from PesaS. In the absence of 3OC6HSL, motA is expressed and motility is restored in the ΔmotA cells. Following addition of 3OC6HSL, EsaR unbinds from PesaS. motA expression decreases and the cells become non-motile. (b) 3OC6HSL gradients were established by adding 0–53 μg of 3OC6HSL on a Whatmann membrane and allowing it to diffuse for 8 h. 10 μM of 3OC6HSL would be the final concentration if 53 µg of 3OC6HSL diffused uniformly through the plate. CoMot-S (ΔmotA transformed with plasmids containing PesaS-motA and Pσ70-esaR) or CoMot-S+ (ΔmotA transformed with plasmids containing PesaS-motA and Pσ70-esaR-D91G) cells were then inoculated at the centre of the plate. Representative plate images obtained after 24 h of incubation at 30 °C. (c) Forward migration distance was measured as the distance between the inoculation point and the visible edge of migration of cells up the signal gradient after 24 h of incubation at 30 °C. Error bars represent one standard deviation from the mean forward migration distance of three biological replicates.
