Figure 1.
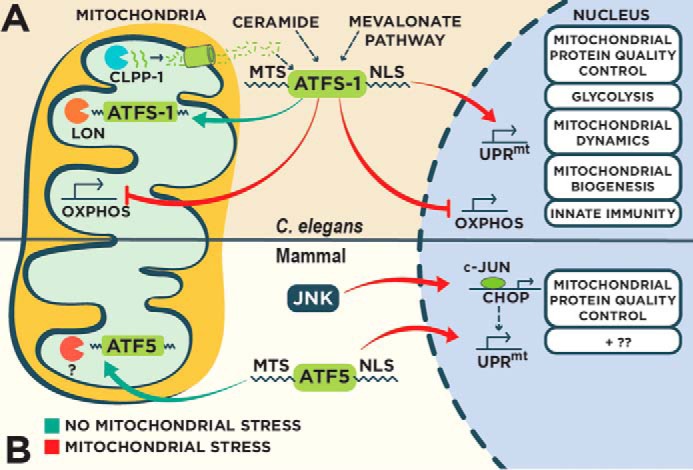
Regulation of the UPRmt in C. elegans and mammals. A, in C. elegans, the UPRmt is principally regulated by the bZIP transcription factor ATFS-1 that contains both mitochondrial and nuclear sorting sequences. In the absence of mitochondrial stress, ATFS-1 localizes to mitochondria and is degraded by the protease Lon. Perturbation to mitochondrial function reduces mitochondrial import efficiency causing an accumulation of cytosolic ATFS-1 and subsequent nuclear import. ATFS-1 regulates a diverse transcriptional program to recover mitochondrial function, including the attenuation of OXPHOS gene expression in both the nucleus and mitochondria. Multiple regulators of the UPRmt have been identified, including the protease ClpP and the ABC transporter HAF-1 that control UPRmt activity through an unidentified mechanism involving peptide efflux. B, mammalian UPRmt is regulated by the transcription factors CHOP and ATF5 that transcriptionally regulate genes to restore mitochondrial homeostasis. CHOP is transcriptionally activated by transcription factor c-Jun during mitochondrial stress. ATF5 contains mitochondrial and nuclear localization sequences similar to ATFS-1 that regulate UPRmt activity based on the efficiency of mitochondrial import.
