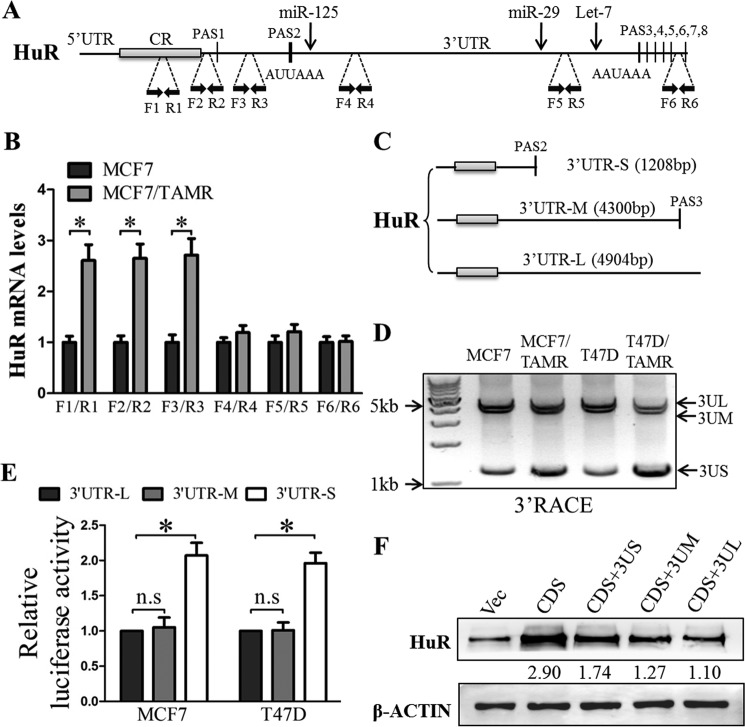
Figure 7.
The 3′-UTRs of HuR transcripts were shortened via APA in TAMR breast cancer cells. A, schematic illustration of HuR mRNA with several PASs in the HuR 3′-UTR. Positions of the primer pairs used in mRNA expression analyses are marked by black dashed lines. Positions of some miRNAs targets are marked by black arrows. B, the levels of HuR mRNA isoforms terminated at different PASs (amplified using the indicated primer pairs) in MCF7 control and TAMR cells were assessed by RT-qPCR analysis. C, schematic illustration of different HuR mRNA isoforms mediated through APA. D, cDNA was synthesized from total RNA isolated from MCF7, MCF7/TAMR, T47D, or T47D/TAMR using anchor oligo(dT) from the 3′-RACE kit (Roche Applied Science). The resulting amplification products were analyzed using agarose gel electrophoresis. E, HuR short, medium, and long 3′-UTRs were cloned downstream of the Renilla luciferase gene in psiCHECK2, and the resulting vectors were transfected into MCF7 and T47D cells. Luciferase reporter activities regulated by the different HuR 3′-UTR isoforms were determined using the Dual-Luciferase assay. F, the HuR coding sequences (CDS) with short, medium, or long 3′-UTRs were cloned into pIRESneo3. The plasmids carrying the corresponding sequences were transfected into MCF7 cells, and HuR protein levels were determined using Western blotting with β-actin as an input control. *, p < 0.05. Error bars, S.E.