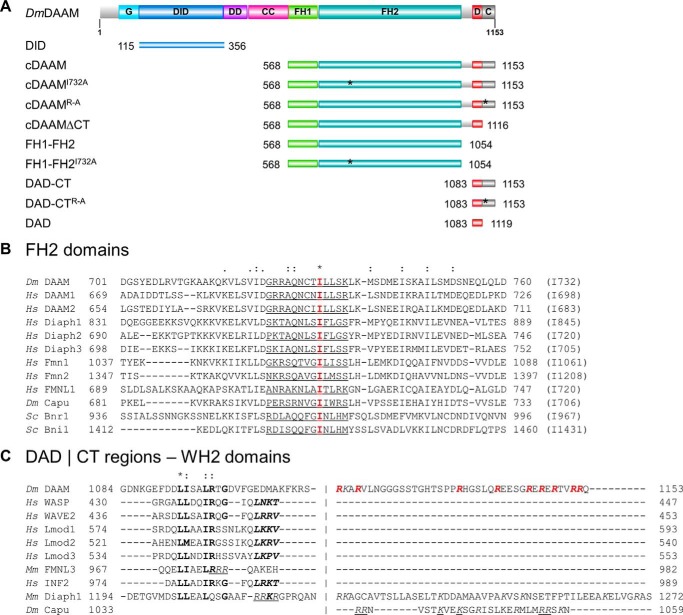
Figure 1.
Domain organization and sequence alignments of DAAM. A, domain organization of full-length Drosophila melanogaster DAAM formin (Dm DAAM) and of the constructs investigated in this study. G, GTPase-binding domain; DID, diaphanous inhibitory domain; DD, dimerization domain; CC, coiled coil; FH, formin homology domain; D, diaphanous autoregulatory domain; CT, C-terminal sequence element. Numbers indicate the number of the first and the last residue in each construct. The positions of the mutated amino acids are highlighted by asterisks. The figure was made with Illustrator for Biological Sciences (57). B, sequence alignment of FH2 domains from different formins. The conserved Ile in the FH2 domain is highlighted in red, and its position in each formin is given in parentheses at the end of the sequences. The αD helix of the knob region is underlined. C, alignment of C-terminal regions from different formins and WH2 domain sequences. The hydrophobic amino acid triplet and the LKK(T/V) motif are shown by bold and bold italics, respectively. Positively charged Arg and Lys residues in the CT region are shown in italics. The Arg residues replaced by Ala in the DAAM DAD-CTArg–Ala construct are highlighted in red. Residues that are shown to be important for actin interaction in mDia1, FMNL3, and Capuccino are underlined (13, 14, 47). UniProt accession numbers are as follows: Dm DAAM, Q8IRY0; Hs DAAM1, Q9Y4D1; Hs DAAM2, Q86T65; Hs Diaph1, O60610; Hs Diaph2, O60879; Hs Diaph3, Q9NSV4; Hs Fmn1, Q68DA7; Hs Fmn2, Q9NZ56; Hs FMNL1, O95466; Dm Capu, Q24120; Sc Bnr1, P40450; Sc Bni1, P41832; Hs WASP, P42768; Hs WAVE2, Q9Y6W5; Hs Lmod1, P29536; Hs Lmod2, Q6P5Q4; Hs Lmod3, Q0VAK6; Mm FMNL3, Q6ZPF4; Hs INF2, Q27J81. Dm, D. melanogaster; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Mm, Mus musculus.