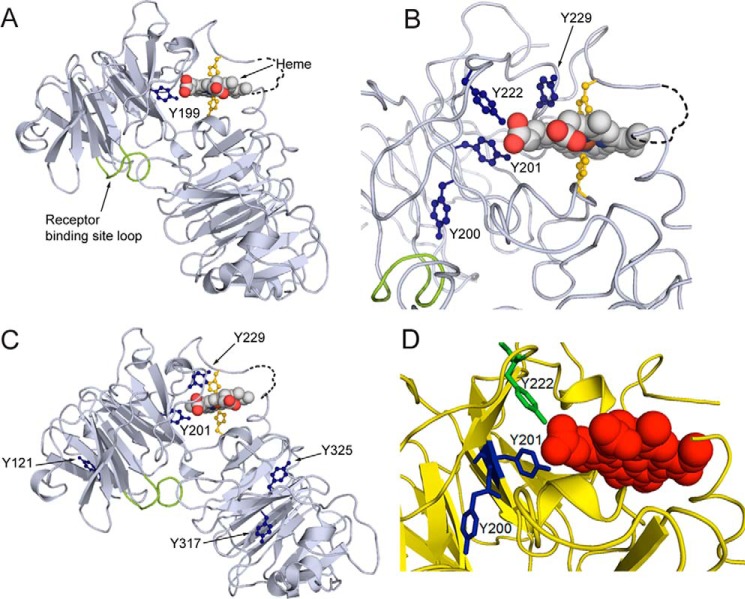
Figure 2.
Covalent oxidatively modified residues modeled on the rabbit hemopexin crystal structure show a cluster of amino acids in the functionally important heme-binding site. A, when the human hemopexin amino acid sequence is threaded on the rabbit hemopexin crystal structure, the nitrated tyrosine Tyrn-199 in the human sequence (labeled and marine blue; equivalent to Tyrn-201 in rabbit, B) resides in the heme-binding site. Nitration of this tyrosine after incubation of hemopexin with N/MPO/GO in vitro is conserved in human, rabbit, and rat proteins. Also shown are the heme-iron coordinating histidine residues (yellow), the receptor binding site (loop only shown of the Jen 14 epitope (lime)) (17), and the linker peptide modeled on the amino acid backbone (dashed black line). B, in the heme-binding site, the endogenously nitrated Tyr-201 and Tyr-222 interact directly with the propionate group on the D ring of the heme and contribute with other aromatic resides to the hydrophobic environment around the heme. Tyr-201 and Tyr-222 help surround this heme propionate together with Arg-199 and Arg-210 from the N-domain and three histidine residues from the C-domain (data not shown). C shows the endogenously nitrated tyrosine residues in rabbit hemopexin identified by LC-MS/MS and summarized in Table 1. D, in this close-up view, Tyr-200 clearly points away from the heme in contrast to Tyr-201 and Tyr-222 (green). These structures were generated using PyMOL (Molecular Graphics System, version 1.3).