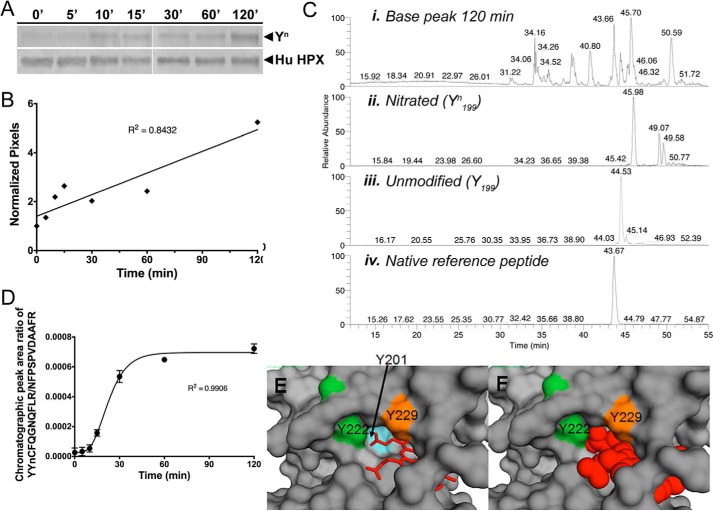
Figure 3.
Time course of in vitro nitration of Tyr-199 in human hemopexin. Time course of in vitro nitration of Tyr-199 in human hemopexin. A and B, Western immunoblotting and quantitation using UN-SCAN-IT software show the relative rate of appearance of nitrotyrosine (Tyrn) in human hemopexin compared with total hemopexin (normalized pixels) after incubation with N/MPO/GO. C, chromatographic peaks from SRM analysis for human hemopexin after 120 min of incubation with N/MPO/GO are shown for the base peak (panel i), nitrated peptide, YYnCFQGNQFLR (panel ii), unmodified peptide, YYCFQGNQFLR (panel iii), and internal reference peptide, NFPSPVDAAFR, for normalizing (panel iv). They were quantified by integration using the native reference peptide method (11). The two minor peaks in the unmodified peptide mass chromatogram that appear to the right of the major peak (panel ii) are unrelated ions with the same m/z, coincidentally. These did not interfere with and were not included in the peak area integration for quantitative analysis. D, time course of nitration using SRM data was quantitated using calibration curves (see supplemental Fig. 4) to determine the mole fraction of in vitro nitrated Tyr-199 in human hemopexin over the period of 120 min. Average peak ratio (nitrated/internal reference) of three separate gel excisions (biological replicates) and analysis per time point is shown ± S.D. Standard curves were calibrated using synthetic peptide chromatographic peak area ratios at each mole fraction point. Experimental chromatographic peak area ratios were obtained by dividing the reference peptide peak area by that of either the YYnCFQGNQFLR (E) or the YYCFQGNQGLR (F) peptide. Mole fractions were calculated as described under “Experimental procedures.” The mole ratio for nitration increased with time of incubation with the N/MPO/GO-nitrating system. E and F show a surface representation of the atoms of rabbit hemopexin with a close-up view of the three tyrosine residues that interact directly with the heme (shown as a stick representation in E and space-filling model in F). Tyr-201 is barely visible when heme is bound, consistent with the vulnerability to oxidative modification of this amino acid residue in the apoprotein. The label for Tyr-222 is positioned over the ring side chain, and this amino acid is protected by a loop (see also Fig. 2D).