Figure 3.
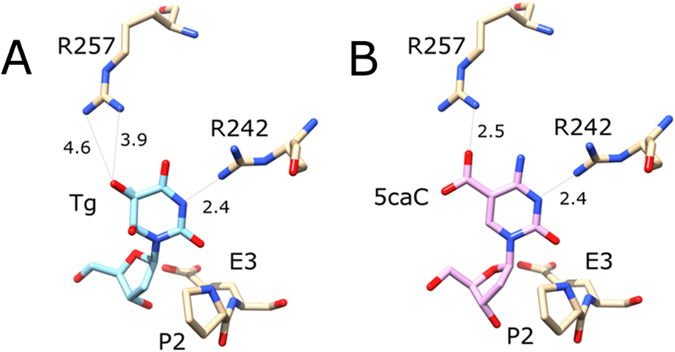
Structural plausibility of the NEIL1 glycosylase activity against DNA containing 5caC: (A) Composite model of the active site with flipped 2′-deoxythymidine glycol. The figure was drawn based on the coordinates determined by Zhu and coworkers for the P2G variant (PDB accession 5ITY)29. The coordinates for the P2 residue in the wild-type were taken from the structure of NEIL1 with a THF abasic site analogue (PDB accession 5ITT)29. Distances in panel A are experimental distances for the molecule A in the structure 5ITY. Both arginine residues adopt different conformations in the other molecules of the asymmetric unit. Note that the pocket for the base is completely obliterated in the 5ITT structure with abasic site analogue. (B) Composite model of the active site with flipped 5caC 2′-deoxynucleotide. The 5caC 2′-deoxynucleotide was taken from the structure of TDG with DNA containing 5caC (PDB accession 3UO7)53, and placed based on least squares superposition of the (planar) 5caC base on the (non-planar) thymidine glycol base. Note that Tg has been reported to adopt a lactim tautomeric structure with hydrogen bond acceptor in the N3 position29. 5caC also has a hydrogen bond acceptor in this position, and is likely protonated in this position to support departure of the base, as suggested for TDG before45. The catalytic residues P2 and E3 are strictly conserved. The position of R242 is subject to RNA editing, making this residue either R or K37. R257 is conserved in many, but not all vertebrates. Numbers indicate distances in Å.
