Figure 1.
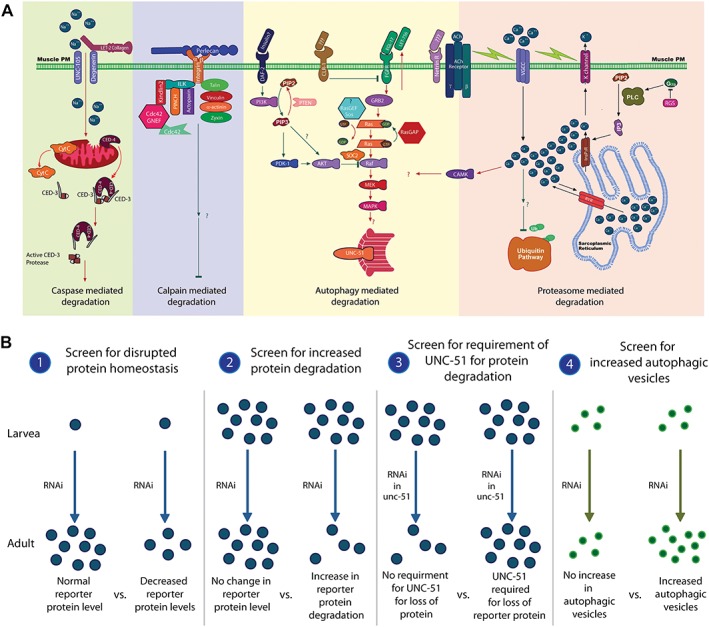
Current model of control of cytosolic muscle protein degradation in Caenorhabditis elegans and schematic of the RNA interference screen for genes potentially regulating autophagy in Caenorhabditis elegans muscle. (A) The model is only from studying the degradation of a single transgenically encoded reporter protein. Far left (green): caspase activation is induced by mitochondrial dysfunction, which can be caused by loss of degenerin channel contact with collagen in the extracellular matrix.19 Left (violet): degradation by calpains is regulated by integrin attachment to the basement membrane,18 and a significant portion of the integrin adhesome appears to contribute to this regulation.16 Middle (yellow): autophagic degradation is controlled by a balance of signal from insulin/insulin‐like receptor (negative regulator, green lines) and autocrine fibroblast growth factor signal (positive regulator, red lines).12, 13, 14 Calcium overload, signalling via CaMKII, also promotes autophagic degradation17 as does knockdown of a number of kinases.20 Right (pink): intracellular calcium controlled by a combination of membrane depolarization, and G‐protein signalling events are required to negatively regulate proteasome‐based degradation.15, 17 Displayed model is adapted from models published in Shephard et al. 17 and Gaffney et al. 19 (B) A schematic of the full RNA interference screen can be found in the kinase screen20 which this phosphatase screen is based upon. Briefly, for identification of phosphatase, genes whose knockdown induced autophagic protein degradation was achieved through four steps: (1) genes for which RNA interference produced decreased amounts of reporter protein in muscle were identified. (2) RNA interference against genes identified in (1) was applied to fully developed adult animals to identify RNA interference treatments that produced degradation of the reporter protein in a muscle. (3) RNA interference against genes identified in (2) was applied to fully developed adult unc‐51 mutant animals to identify RNA interference treatments that failed to produce degradation in the absence of functional UNC‐51. (4) RNA interference against genes identified in (3) was applied to fully developed adult animals containing GFP tagged LGG‐1 to identify RNA interference treatments that produced elevated levels of autophagic vesicles.
