Abstract
Crop productivity needs to substantially increase to meet global food and feed demand for a rapidly growing world population. Agricultural technology developers are pursuing a variety of approaches based on both traditional technologies such as genetic improvement, pest control and mechanization as well as new technologies such as genomics, gene manipulation and environmental modelling to develop crops that are capable of meeting growing demand. Photosynthesis is a key biochemical process that, many suggest, is not yet optimized for industrial agriculture or the modern global environment. We are interested in identifying control points in maize photoassimilation that are amenable to gene manipulation to improve overall productivity. Our approach encompasses: developing and using novel gene discovery techniques, translating our discoveries into traits and evaluating each trait in a stepwise manner that reflects a modern production environment. Our aim is to provide step change advancement in overall crop productivity and deliver this new technology into the hands of growers.
This article is part of the themed issue ‘Enhancing photosynthesis in crop plants: targets for improvement’.
Keywords: C4 photosynthesis, lead discovery, agricultural biotechnology, mathematical modelling, gene regulatory network
1. Background
To meet projected demand for food, fibre and energy, crop productivity needs to continue on an upward trajectory. The world population is expected to reach 9 billion by 2050 which will require food production to nearly double to maintain today's standards. Current trends suggest that productivity improvements for the world's four major crops will fall well short of this goal [1]. Agricultural producers draw on a multitude of technologies to ensure efficient, sustainable, stable and high-quality crop production. Genetics has been the foundation of crop improvement since the dawn of agriculture. New genome technologies have transformed genetics into an information-rich (driven) discipline.
Photosynthesis is a viable plant productivity improvement target [2–5]. Since its discovery [6], C4 photosynthesis has become widely recognized as an effective photosynthetic mechanism for hot, dry climates, increasing both nitrogen and water use efficiency [7]. The desire to transfer the C mechanism to less efficient C3 crops such as rice [8,9] brought new attention and funding to C4 research. This has produced new tools to support C4 photosynthesis research including development of model species to investigate genetics [10], and tools to support genome-wide analysis of the underlying biology [11]. Several comparative studies are underway to determine requirements to introduce C4 metabolism into C3 crops [12,13].
This renewed interest in C4 metabolism has led to advances in genomics [14–17] and metabolomics [18] that have expanded the C4 network. For example, it is now understood that crops such as maize belong to an NADP–malic enzyme (NADP-ME) type, with two CO2 pumps to facilitate CO2 concentration around Rubisco in different environments [19,20]. The properties of enzymes involved in C4 metabolism are being characterized in detail [21–24]. Investigators are also examining the role of transport processes between the mesophyll and bundle sheath [25], regulation of C4 metabolism [18] and photoprotection [26,27]. Computer modelling is also advancing knowledge of C4 metabolism [28,29]. At the turn of the millennium, there was sufficient knowledge to construct a fairly accurate mathematical model of the NADP-ME C4 process [30]. This model was recently updated [17]. All this activity has greatly improved the understanding of C4 metabolism.
Genetic engineering (GM) technology proved transformative, becoming the most rapidly adopted crop improvement technology in history [31]. Its principal applications are in insect and weed control. Goals that target intrinsic biological processes have been less accessible to manipulation by GM technology. However, some promising advancements have emerged [32–34]. GM technology is most effective when metabolic control points [35] are targeted. These are enzymes or regulatory factors that significantly influence the biological process of interest. They are notoriously difficult to identify [36]. The agricultural biotechnology discovery process aims to efficiently and effectively identify candidate genes, or leads, that fit this definition. Tools developed for this include a variety of genetic approaches such as TILLING, transposon tagging and forward or reverse gene tagging [37,38]. Each has proved effective in a number of plant species but can be expensive, and perhaps intractable, when targeting a complex metabolic process like C4 photosynthesis, because there are few robust methods to measure photosynthetic traits. Now many organizations are turning to computational tools as entry points for lead discovery [39,40].
Genomics makes it possible to execute information-driven lead discovery strategies [41]. Not all crops are accessible to this approach, because the ideal starting point is a high-quality genome sequence. A genome sequence enables layers of genetic information, such as gene annotation, RNA transcripts, protein-coding sequence, metabolites, genetic markers and chromatin data, to then be applied across the genome. Several genome-wide analytical tools have been developed to interrogate these data including genome-wide association analysis, gene regulatory networks and mathematical modelling of metabolic pathways. These tools make it possible to identify genes that are less accessible to genetic screens [42,43]. For example, a genetic lesion may not express at the physiological level but might be easily detected at the gene level. These tools have also been applied to metabolic processes [44].
This work aims to identify genes predicted to improve C4 photosynthesis using GM technology, and which represent leads for trait development. It began with a mathematical model of the NADP-ME-type C4 biochemical network [30] that could be interrogated for enzymes predicted to exert significant control over photoassimilation. To expand this effort, a maize gene regulatory network was constructed to identify genes predicted to regulate C4 metabolism. More recently, genome-wide association tools became available as the primary and secondary lead sources. As computational tools are at least one step removed from direct plant observation, data quality and accuracy need to be assessed using independent evidence. The objective is to compare the weight of evidence for each lead identified by a computational process. This prioritizes leads for GM trait development, and can also form the basis for breeding or genome-editing strategies. We focused on maize applications and developed strategies to identify leads that might be amenable to GM manipulation. While each area defined a focal point, none were pursued in isolation. High-quality leads were defined as those with support from multiple types of evidence. Information-driven lead identification for the purpose of directly enhancing crop performance mechanisms is a relatively new and unproven approach. The purpose of this report is to illustrate the application of modern computational tools such as genome-wide association studies (GWAS), gene regulatory network analysis and mathematical modelling to prioritize candidate leads for manipulation through genetic modification. A prioritized set of leads can then be evaluated in an industrial trait development pipeline for increased field performance.
2. Genome-wide association analysis
As a first step to identify leads of importance to photosynthetic improvement, we took advantage of a dataset generated previously that evaluated a range of maize germplasms for dry grind ethanol production. Several traits described in electronic supplementary material, table S1 are key attributes for maize starch as an ethanol substrate that overlap with plant yield characteristics and have potential as a proxy for productivity or yield. Starch represents approximately 70% of grain weight, and as such is considered a yield characteristic for both ethanol production and overall plant performance. More importantly, it was an excellent dataset for genome-wide association analysis, because there was high replication and large variation in the recorded traits. These characteristics often increase the power of association mapping to detect genomic regions linked to trait variation. Furthermore, the recorded phenotypes exhibited a normal distribution, which satisfies a critical assumption of association mapping, and some phenotypes correlated with each other. Quality phenotypic data were collected across 493 maize inbred lines for per cent ash (ASH_P), kernel density (DEN_N), dry grind ethanol produced after 24, 48 and 72 h fermentation (i.e. DG24P), grain moisture, percentages of oil, protein, starch and crude fibre.
High-throughput sequence analysis of the transcriptome (mRNA-Seq) for approximately 380 of these lines was used for single nucleotide polymorphism (SNP) discovery. SNP tagging analysis was performed to create a set of non-redundant SNPs for downstream genetic diversity and association analysis. Often, nearby SNPs are inherited together leading to high correlations of alleles among nearby SNPs, so one SNP was selected to represent a group of SNPs in high linkage disequilibrium, at a correlation R2 threshold of 80%. A minor allele frequency (MAF) filter at 0.01 was also performed, to avoid misleading results due to small sample sizes resulting from rare alleles. The 0.01 threshold was chosen to select SNPs whose rare allele occurred in at least four lines in the panel. After these filters, approximately 140 000 SNPs were retained. The tagged SNP dataset was then used to create a pairwise kinship matrix using a simple proportion of identical homozygous genotypes. This matrix quantifies the degree of relationship of the panel lines in a pairwise manner and was used to control for population structure in association mapping.
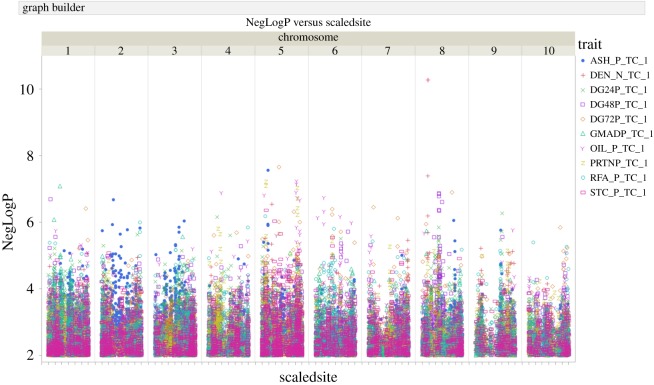
Associations between SNPs and phenotypes were detected using Tassel 4.0 [45]. The best linear unbiased predictor (BLUP) values for the 10 traits, the kinship matrix based on the tagged SNP dataset and the tagged SNP genotype dataset were used as input files for the association mapping using the Mixed Linear Models approach implemented in Tassel 4.0 [45]. The Manhattan plot in figure 1 shows the results across the 10 chromosomes of maize. Although a negative log P (NegLogP) > 3 is significant, a more stringent threshold was set at NegLogP > 6 to identify 22 genes of interest (table 1) that qualify as leads for further analysis.
Figure 1.
Manhattan plot of dry grind ethanol traits in a proprietary inbred maize population. The significance of associations is expressed as the negative base 10 log of the p-value, on the Y-axis. The positions of associations within each of the 10 chromosomes are indicated by the X-axis. Each point in the plot represents results of association between one polymorphism and one trait. Traits are indicated by different shapes and symbols as shown on the upper right of the plot diagram.
Table 1.
Leads identified by GWAS analysis of maize dry grind ethanol traits. The chromosome location of each lead is based on the maize V2 genome. The traits are described in electronic supplementary material, table S1.
| Gramene ID | linkage group | trait | description | begin | end | F | p-value | NegLogP |
|---|---|---|---|---|---|---|---|---|
| GRMZM2G012874 | 8 | DG72P | beta3-glucuronyltransferase, uncharacterized protein | 131688249 | 131694614 | 28.9817 | 1.29 × 10−7 | 6.8903 |
| GRMZM2G029912 | 9 | DG24P | uncharacterized protein | 105190710 | 105197290 | 25.9833 | 5.46 × 10−7 | 6.2628 |
| GRMZM2G034302 | 1 | DG48P | uncharacterized protein | 15069803 | 15075192 | 28.0265 | 2.04 × 10−7 | 6.6911 |
| GRMZM2G035741 | 6 | OIL_P | uncharacterized protein | 35892952 | 35896625 | 26.2301 | 4.85 × 10−7 | 6.3147 |
| GRMZM2G056600 | 7 | DG72P | uncharacterized protein | 129263776 | 129265926 | 25.2905 | 7.64 × 10−7 | 6.1168 |
| GRMZM2G066460 | 6 | OIL_P | 60S acidic ribosomal protein P0 | 9203729 | 9206286 | 25.3335 | 7.48 × 10−7 | 6.1259 |
| GRMZM2G074604 | 5 | OIL_P | phenylalanine ammonia-lyase | 186677004 | 186680745 | 30.1549 | 7.35 × 10−8 | 7.134 |
| GRMZM2G089454 | 5 | OIL_P | uncharacterized protein | 204908277 | 204911697 | 27.8408 | 2.23 × 10−7 | 6.6523 |
| GRMZM2G097884 | 5 | OIL_P | uncharacterized protein | 190953138 | 190954236 | 28.171 | 1.90 × 10−7 | 6.7212 |
| GRMZM2G108474 | 5 | STC_P | translationally controlled tumour protein homologue | 68020016 | 68022909 | 24.8886 | 9.29 × 10−7 | 6.0319 |
| GRMZM2G114127 | 5 | OIL_P | uncharacterized protein | 184393704 | 184395078 | 28.9527 | 1.31 × 10−7 | 6.8842 |
| GRMZM2G122076 | 4 | OIL_P | putative homeobox DNA-binding and leucine zipper domain family protein | 77609764 | 77614223 | 28.8919 | 1.34 × 10−7 | 6.8716 |
| GRMZM2G137930 | 6 | OIL_P | soluble inorganic pyrophosphatase, uncharacterized protein | 35680626 | 35684863 | 28.1908 | 1.88 × 10−7 | 6.7254 |
| GRMZM2G142093 | 5 | DEN_N | uncharacterized protein | 53179092 | 53180352 | 27.3245 | 2.86 × 10−7 | 6.5443 |
| GRMZM2G145573 | 8 | DG48P | catalytic/oxidoreductase acting on NADH or NADPH | 87238606 | 87243400 | 27.6341 | 2.46 × 10−7 | 6.6091 |
| GRMZM2G152041 | 6 | DG72P | uncharacterized protein | 71014229 | 71016297 | 25.8101 | 5.94 × 10−7 | 6.2263 |
| GRMZM2G152764 | 5 | RFA_P | no description | 18698735 | 18701561 | 25.5729 | 6.66 × 10−7 | 6.1763 |
| GRMZM2G155662 | 6 | DG72P | single-stranded DNA-binding protein WHY1, chloroplastic | 71618436 | 71621497 | 26.5803 | 4.09 × 10−7 | 6.3882 |
| GRMZM2G163296 | 1 | DG72P | putative uncharacterized protein | 283309246 | 283311287 | 26.6681 | 3.92 × 10−7 | 6.4067 |
| GRMZM2G169114 | 6 | OIL_P | uncharacterized protein | 104848940 | 104858629 | 25.4782 | 6.98 × 10−7 | 6.1564 |
| GRMZM2G431900 | 7 | DG72P | uncharacterized protein | 22154867 | 22161180 | 26.8555 | 3.58 × 10−7 | 6.446 |
| GRMZM2G540538 | 5 | OIL_P | adenosine kinase 2, uncharacterized protein | 186517098 | 186522957 | 30.5717 | 6.02 × 10−8 | 7.2203 |
3. Gene regulatory network analysis
Gene regulatory network analysis seeks to identify relationships between components of complex datasets. In this case, mRNA profiling data were used to find key regulators of C4 metabolism. Network inference algorithms were used to generate C4 photosynthesis leads based on hypotheses in the form element X regulates element Y under W conditions. A number of gene regulatory relationships were established using gene expression data. The algorithms came from the dialogue for reverse engineering assessments and methods (DREAM) challenge. This systems biology approach has been used to identify important master regulators of high-grade gliomas [46], characterize carbon and nitrogen gene regulation in Arabidopsis [47] and identify Staphylococcus aureus transcription factors involved in pathogenesis [48]. However, no single algorithm can accurately infer all network motifs, and some algorithms are complementary [48]. As such, several algorithms were reviewed and combined to analyse and evaluate gene regulation of the C4 photosynthesis network.
First, microarray data from four studies representing 216 maize leaf samples were used to develop a network. Data were generated as previously described [49]. To ensure robustness, samples below a predetermined quality score were removed, ambiguously labelled probe sets were excluded and remaining probe sets were filtered based on negative controls. A total of 16 179 genes were measured. The algorithms incorporated into the analysis included TIGRESS [50], C3NET [51], Pearson correlation [40] and GENIE3 [52]. The default parameters of each algorithm were used to predict gene interactions. A consensus network was then created from the networks generated by each method as described in Marbach et al. [48].
A second dataset was created by focusing on genes annotated as transcription factors. Transcription factors are good candidates for control points and master regulators of plant metabolism due to their regulatory role, controlling the expression of metabolic genes. A set of maize transcription factors was identified from three sources: Grassius [53], PlantTFDB [54] and Gene Ontology [55,56]. Master regulators were then determined from this set based on the connections of the transcription factors with C4 pathway genes in the consensus network via Fisher's Exact Test [46]. These master regulators were predicted to significantly control C4 photosynthesis. Core C4 genes formed the basis of the analysis. The master regulator method defines a statistically significant relationship between gene interactions. It identifies transcription factors predicted to regulate several genes in the network and identifies genes that are potential lead candidates.
Independently each gene network method produces many false positives and these false positives carry through into the consensus network. Thus, a final filtering step was performed to create a focused list of genes for further investigation. A number of multiple linear regression models that predicted the combination of connected genes that can predict the expression of the genes involved in the C4 network were tested. Only the models with the smallest standard error were retained. This final set then formed the final gene network. Not all master regulators survived this final filtering step. There was overlap with the GWAS results, the master regulator analysis and multiple linear regression models which identified the genes in table 2 as putative regulators of C4 metabolism.
Table 2.
Leads identified by gene regulatory network analysis of maize leaf gene expression profiling data. The chromosome location of each lead is based on the maize V2 genome.
| Gramene ID | description | linkage group | start | end |
|---|---|---|---|---|
| GRMZM2G026024 | uncharacterized protein | 4 | 158996579 | 158993661 |
| AC204212.4_FG001 | SANT/MYB protein | 5 | 201120649 | 201120939 |
| GRMZM2G017429 | uncharacterized protein | 5 | 207899002 | 207900503 |
| GRMZM2G440529 | putative uncharacterized protein | 3 | 190314296 | 190317061 |
| GRMZM2G372102 | 36.4 kDa proline-rich protein | 2 | 5922079 | 5922834 |
| GRMZM2G306732 | fructose-1,6-bisphosphatase | 1 | 38850785 | 38848595 |
| GRMZM5G886257 | malic enzyme | 6 | 130130465 | 130134088 |
| GRMZM2G456568 | putative NAC domain transcription factor superfamily protein isoforms 1 and 2 | 6 | 147925808 | 147921021 |
| GRMZM2G113779 | no description | 2 | 201972947 | 201971116 |
| GRMZM5G863645 | uncharacterized protein | 1 | 269289476 | 269284325 |
| GRMZM2G060265 | no description | 1 | 1955584 | 1957660 |
4. Mathematical modelling of C4 photoassimilation
Mathematical models can be powerful tools to identify control points in a metabolic network. A mathematical model of C4 photoassimilation [30] was used as the basis for analysing the contribution of each enzyme (or network component) to photoassimilation. An in silico transformation algorithm was developed to interrogate the network for control points that might increase photoassimilation if enzyme activity were increased. Given the thousands of possible gene combinations, a molecular genetic or empirical approach to this problem is intractable. Model interrogation was used to narrow the solutions to those most likely to have a positive impact. It also revealed the theoretical maximum improvement that might be achievable.
Upregulation at each enzyme step was examined alone and in combination with other steps. First, a systematic analysis identified 1, 2, 3 and 4 component combinations that increased assimilation by a minimum of 5%. This narrowed the solutions an order of magnitude from thousands to hundreds. Next, the sensitivity of photoassimilation to the model's predicted level of upregulation was examined. This was necessary because genetic engineering is not an exact science. It is impossible to predict exactly how a trait gene will function once stably integrated into the host genome. Furthermore, the most viable solutions involve three and four genes which will likely influence trait behaviour in ways that have not been explored in much detail. This further reduced the solutions to tens. Finally, the frequency a particular enzyme appeared in a solution was tabulated. Table 3 shows that NADP-ME topped the list followed by phosphoenolpyruvate carboxylase, fructose-bisphosphate aldolase and NADP-malate dehydrogenase.
Table 3.
Enzymes most often found as leads in mathematical modelling solutions that are predicted to increase photoassimilation by at least 5%. The enzyme information is from the BRENDA database (http://www.brenda-enzymes.org/). The enzymes are ranked based on the number of times they occur in a mathematical modelling solution. Four different scenarios are represented.
| name of enzyme or enzyme complex | common name | EC number | cell type | subcellular localization | occurrences as a single gene solution | occurrences in 2-gene candidate combinations | occurrences in 3-gene candidate combinations | occurrences in 4-gene candidate combinations | total occurrences in all gene candidate combinations |
|---|---|---|---|---|---|---|---|---|---|
| malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) | NADP-malic enzyme | 1.1.1.40 | bundle sheath | chloroplast | 1 | 13 | 67 | 214 | 295 |
| phosphoenolpyruvate carboxylase | PEPC | 4.1.1.31 | mesophyll | 0 | 5 | 42 | 173 | 220 | |
| fructose-bisphosphatase | FBPase | 3.1.3.11 | bundle sheath | chloroplast | 0 | 3 | 31 | 156 | 190 |
| malate dehydrogenase (NADP+) | NADP-malate dehydrogenase | 1.1.1.82 | mesophyll | 0 | 2 | 27 | 136 | 165 | |
| fructose-bisphosphate aldolase | aldolase | 4.1.2.13 | bundle sheath | chloroplast | 0 | 3 | 27 | 124 | 154 |
| 6-phosphofructo-2-kinase | fructose 2,6-bisphosphate aldolase | 2.7.1.105 | bundle sheath | cytosol | 0 | 2 | 23 | 117 | 142 |
| phosphoribulokinase | phosphoribulokinase | 2.7.1.19 | bundle sheath | chloroplast | 0 | 2 | 21 | 112 | 135 |
| pyruvate, phosphate dikinase | PPDK | 2.7.9.1 | mesophyll | 0 | 1 | 17 | 111 | 129 | |
| sedoheptulose-bisphosphatase | SBPase | 3.1.3.37 | bundle sheath | chloroplast | 0 | 1 | 18 | 109 | 128 |
| phosphoglycerate kinase/ glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) | PGA kinase/GAP dehydrogenase complex | 2.7.2.3/1.2.1.13 | bundle sheath | chloroplast | 0 | 1 | 18 | 107 | 126 |
| phosphoglycerate kinase/ glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) | PGA kinase/GAP dehydrogenase complex | 2.7.2.3/ 1.2.1.13 | mesophyll | 0 | 2 | 21 | 96 | 119 |
There were no single or double enzyme combinations that met established performance criteria to improve photoassimilation by at least 5%. This suggests shared control of overall network flux. It is notable that no further improvement was predicted when more than four enzymes were manipulated. Some steps such as ribulose-bisphosphate carboxylation were not examined because perturbation at that step is not accessible to current genetic engineering technology. Ribulose-bis-phosphate carboxylase/oxygenase (Rubisco), the enzyme that catalyses this step, is a heterooctamer consisting of a large subunit encoded in the plastid genome and a small subunit encoded by a three-member gene family in the nuclear genome. Since the holoenzyme has subunits from both nuclear and chloroplast compartments, there is no simple way to upregulate its expression. Electronic supplementary material, table S2 shows that the top three-gene solutions are predicted to improve photoassimilation by just over 11%, and electronic supplementary material, table S3 shows the top four-gene solutions are predicted to produce just over 13% improvement.
5. Prioritizing computational leads for trait development
The computational approaches listed above provide compelling evidence implicating new genes as control points in C4 carbon assimilation. However, several additional steps are required to translate gene leads derived from computational approaches into targets for transgenic genetic modification. First, computational discovery methodology relies heavily on maize genome annotation which remains relatively incomplete relative to model plants. While robust leads are often identified by more than one approach, many of the leads presented here are often genes that have not been previously investigated. Additional evidence helps to build confidence in a lead and prioritize it for further investigation. Once identified, each lead was analysed using a variety of tools and resources. For example, internal databases show if a lead is associated with yield quantitative trait loci or other genetic characteristics that might be of interest. A lead's expression pattern can inform on its role in photoassimilation as well as other biological processes. These data can also be used to determine if a lead responds to environmental signals such as temperature, water availability or nitrogen. The protein-coding sequence may reveal functional information that can inform on the mechanism by which the lead influences photoassimilation. It is also useful to know if a lead has been subject to transgenic experiments or if genetic mutants at that locus have been characterized. In some cases, a lead's orthologue in another species has been studied in detail, and can reveal a specific functional role. Taken together, a rich body of information can be applied that formulates a compelling hypothesis and is supported by the available evidence consistent with trait development objectives.
While most of the strategy focus is aimed at GM technology as a solution for increased photosynthetic efficiency, leads identified by computational methods can also be evaluated as markers for targeted breeding. This provides an alternative to GM technology products which are challenging and costly to commercialize due to the rigorous health and safety assessments required for registration. Of course, a breeding approach is limited to current genetic variation and does not take advantage of the broader genetic manipulations possible through GM technology.
6. Concluding remarks
Genetic modifications through biotechnology are powerful crop improvement tools. However, GM has been less successful with respect to improving intrinsic plant processes important to plant performance traits, such as photosynthesis. Theory suggests that enhancing C4 carbon assimilation is one way to improve crop productivity. Much is known about the C4 process, and recent work suggests there is much more to learn. Maize genomics has greatly expanded trait development opportunities. To take advantage of this, we developed computational tools to scan the genome for high-quality trait development leads. This represents the next wave in lead discovery and will pave the way to new high-performing crops to meet the needs of a growing global population.
Supplementary Material
Acknowledgements
The authors thank our internal stakeholders who helped shape this work, in particular Kimberly White and Robert Bensen. We acknowledge the support from Syngenta's Performance Plant Biology Division led by Ian Jepson.
Data accessibility
This article has no additional data.
Authors' contributions
M.L.N. and N.J.B. led the team that conceived and conducted the work. S.M.S. designed and carried out the gene regulatory network analysis. J.C. designed and carried out the GWAS analysis. L.P. designed and carried out the mathematical modelling. J.Co., P.E.W., X.T., J.D., J.N., J.T. and R.H. participated in data analysis. M.L.N. wrote the paper.
Competing interests
This work was done at Syngenta Crop Protection, LLC. Syngenta is a private company engaged in the development of agricultural technology products. Some of the work presented in this article is covered by one or more patent applications filed in jurisdictions around the world.
Funding
All work was paid for by Syngenta.
References
- 1.Ray DK, Mueller ND, West PC, Foley JA. 2013. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8, e66428 ( 10.1371/journal.pone.0066428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu XG, Long SP, Ort DR. 2010. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61, 235–261. ( 10.1146/annurev-arplant-042809-112206) [DOI] [PubMed] [Google Scholar]
- 3.Evans JR. 2013. Improving photosynthesis. Plant Physiol. 162, 1780–1793. ( 10.1104/pp.113.219006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ort DR, et al. 2015. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl Acad. Sci. USA 112, 8529–8536. ( 10.1073/pnas.1424031112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long SP, Marshall-Colon A, Zhu XG. 2015. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66. ( 10.1016/j.cell.2015.03.019) [DOI] [PubMed] [Google Scholar]
- 6.Hatch MD. 2002. C4 photosynthesis: discovery and resolution. Photosynth. Res. 73, 251–256. ( 10.1023/A:1020471718805) [DOI] [PubMed] [Google Scholar]
- 7.Hibberd JM, Covshoff S. 2010. The regulation of gene expression required for C4 photosynthesis. Annu. Rev. Plant Biol. 61, 181–207. ( 10.1146/annurev-arplant-042809-112238) [DOI] [PubMed] [Google Scholar]
- 8.Leegood RC. 2002. C4 photosynthesis: principles of CO2 concentration and prospects for its introduction into C3 plants. J. Exp. Bot. 53, 581–590. ( 10.1093/jexbot/53.369.581) [DOI] [PubMed] [Google Scholar]
- 9.Langdale JA. 2011. C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell 23, 3879–3892. ( 10.1105/tpc.111.092098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brutnell TP, Wang L, Swartwood K, Goldschmidt A, Jackson D, Zhu X-G, Kellogg E, Van Eck J. 2010. Setaria viridis: a model for C4 photosynthesis. Plant Cell 22, 2537–2544. ( 10.1105/tpc.110.075309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J, Li Y, Ma X, Ding J, Wang K, Wang S, Tian Y, Zhang H, Zhu XG. 2013. Whole transcriptome analysis using next-generation sequencing of model species Setaria viridis to support C4 photosynthesis research. Plant Mol. Biol. 83, 77–87. ( 10.1007/s11103-013-0025-4) [DOI] [PubMed] [Google Scholar]
- 12.Brautigam A, Hoffmann-Benning S, Weber AP. 2008. Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes. Plant Physiol. 148, 568–579. ( 10.1104/pp.108.121012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, et al. 2014. Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nat. Biotechnol. 32, 1158–1165. ( 10.1038/nbt.3019) [DOI] [PubMed] [Google Scholar]
- 14.Chang YM, et al. 2012. Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 160, 165–177. ( 10.1104/pp.112.203810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P, Kelly S, Fouracre JP, Langdale JA. 2013. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. Plant J. 75, 656–670. ( 10.1111/tpj.12229) [DOI] [PubMed] [Google Scholar]
- 16.Brautigam A, Schliesky S, Kulahoglu C, Osborne CP, Weber AP. 2014. Towards an integrative model of C4 photosynthetic subtypes: insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. J. Exp. Bot. 65, 3579–3593. ( 10.1093/jxb/eru100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Long SP, Zhu XG. 2014. Elements required for an efficient NADP-malic enzyme type C4 photosynthesis. Plant Physiol. 164, 2231–2246. ( 10.1104/pp.113.230284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pick TR, et al. 2011. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell 23, 4208–4220. ( 10.1105/tpc.111.090324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellasio C, Griffiths H. 2014. The operation of two decarboxylases, transamination, and partitioning of C4 metabolic processes between mesophyll and bundle sheath cells allows light capture to be balanced for the maize C4 pathway. Plant Physiol. 164, 466–480. ( 10.1104/pp.113.228221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Brautigam A, Weber AP, Zhu XG. 2014. Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. J. Exp. Bot. 65, 3567–3578. ( 10.1093/jxb/eru058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Von Caemmerer S, Quinn V, Hancock NC, Price GD, Furbank RT, Ludwig M. 2004. Carbonic anhydrase and C4 photosynthesis: a transgenic analysis. Plant Cell Environ. 27, 697–703. ( 10.1111/j.1365-3040.2003.01157.x) [DOI] [Google Scholar]
- 22.Wang D, Portis AR Jr, Moose SP, Long SP. 2008. Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold tolerance of carbon assimilation in Miscanthus x giganteus. Plant Physiol. 148, 557–567. ( 10.1104/pp.108.120709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarez CE, Saigo M, Margarit E, Andreo CS, Drincovich MF. 2013. Kinetics and functional diversity among the five members of the NADP-malic enzyme family from Zea mays, a C4 species. Photosynth. Res. 115, 65–80. ( 10.1007/s11120-013-9839-9) [DOI] [PubMed] [Google Scholar]
- 24.Paulus JK, Schlieper D, Groth G. 2013. Greater efficiency of photosynthetic carbon fixation due to single amino-acid substitution. Nat. Commun. 4, 1518 ( 10.1038/ncomms2504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brautigam A, Weber APM. 2011. Do metabolite transport processes limit photosynthesis? Plant Physiol. 155, 43–48. ( 10.1104/pp.110.164970) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubbart S, Ajigboye OO, Horton P, Murchie EH. 2012. The photoprotective protein PsbS exerts control over CO2 assimilation rate in fluctuating light in rice. Plant J. 71, 402–412. ( 10.1111/j.1365-313X.2012.04995.x) [DOI] [PubMed] [Google Scholar]
- 27.Murchie EH, Niyogi KK. 2011. Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol. 155, 86–92. ( 10.1104/pp.110.168831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G-R, Wang Q-F, Zhuang J. 2004. Modeling the water use efficiency of soybean and maize plants under environmental stresses: application of a synthetic model of photosynthesis-transpiration based on stomatal behavior. J. Plant Physiol. 161, 303–318. ( 10.1078/0176-1617-00972) [DOI] [PubMed] [Google Scholar]
- 29.Yin X, Struik PC. 2012. Mathematical review of the energy transduction stoichiometries of C4 leaf photosynthesis under limiting light. Plant Cell Environ. 35, 1299–1312. ( 10.1111/j.1365-3040.2012.02490.x) [DOI] [PubMed] [Google Scholar]
- 30.Laisk A, Edwards GE. 2000. A mathematical model of C4 photosynthesis: the mechanism of concentrating CO2 in NADP-malic enzyme type species. Photosynth. Res. 66, 199–224. ( 10.1023/A:1010695402963) [DOI] [PubMed] [Google Scholar]
- 31.James C. 2010. A global overview of biotech (GM) crops: adoption, impact and future prospects. GM Crops 1, 8–12. ( 10.4161/gmcr.1.1.9756) [DOI] [PubMed] [Google Scholar]
- 32.Castiglioni P, et al. 2008. Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 147, 446–455. ( 10.1104/pp.108.118828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Habben JE, et al. 2014. Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought-stress conditions. Plant Biotechnol. J. 12, 685–693. ( 10.1111/pbi.12172) [DOI] [PubMed] [Google Scholar]
- 34.Nuccio ML, et al. 2015. Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotech. 33, 862–869. ( 10.1038/nbt.3277) [DOI] [PubMed] [Google Scholar]
- 35.Stephanopoulos G. 1994. Metabolic engineering. Curr. Opin. Biotechnol. 5, 196–200. ( 10.1016/S0958-1669(05)80036-9) [DOI] [PubMed] [Google Scholar]
- 36.Yadav VG, De Mey M, Lim CG, Ajikumar PK, Stephanopoulos G. 2012. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab. Eng. 14, 233–241. ( 10.1016/j.ymben.2012.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Candela H, Hake S. 2008. The art and design of genetic screens: maize. Nat. Rev. Genet. 9, 192–203. ( 10.1038/nrg2291) [DOI] [PubMed] [Google Scholar]
- 38.Alonso JM, Ecker JR. 2006. Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat. Rev. Genet. 7, 524–536. ( 10.1038/nrg1893) [DOI] [PubMed] [Google Scholar]
- 39.Huang P, Brutnell TP. 2016. A synthesis of transcriptomic surveys to dissect the genetic basis of C4 photosynthesis. Curr. Opin. Plant Biol. 31, 91–99. ( 10.1016/j.pbi.2016.03.014) [DOI] [PubMed] [Google Scholar]
- 40.Pyzer-Knapp EO, Suh C, Gómez-Bombarelli R, Aguilera-Iparraguirre J, Aspuru-Guzik A. 2015. What Is high-throughput virtual screening? A perspective from organic materials discovery. Annu. Rev. Mater. Res. 45, 195–216. ( 10.1146/annurev-matsci-070214-020823) [DOI] [Google Scholar]
- 41.Chen H, He H, Zhou F, Yu H, Deng XW. 2013. Development of genomics-based genotyping platforms and their applications in rice breeding. Curr. Opin Plant Biol. 16, 247–254. ( 10.1016/j.pbi.2013.04.002) [DOI] [PubMed] [Google Scholar]
- 42.Hirai MY, et al. 2007. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl Acad. Sci. USA 104, 6478–6483. ( 10.1073/pnas.0611629104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhosale R, et al. 2013. Predicting gene function from uncontrolled expression variation among individual wild-type Arabidopsis plants. Plant Cell 25, 2865–2877. ( 10.1105/tpc.113.112268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maruyama K, et al. 2009. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150, 1972–1980. ( 10.1104/pp.109.135327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. ( 10.1093/bioinformatics/btm308) [DOI] [PubMed] [Google Scholar]
- 46.Carro MS, et al. 2010. The transcriptional network for mesenchymal transformation of brain tumours. Nature 463, 318–325. ( 10.1038/nature08712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutierrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. 2007. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 8, R7 ( 10.1186/gb-2007-8-1-r7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marbach D, et al. 2012. Wisdom of crowds for robust gene network inference. Nat. Methods 9, 796–804. ( 10.1038/nmeth.2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coneva V, Zhu T, Colasanti J. 2007. Expression differences between normal and indeterminate1 maize suggest downstream targets of ID1, a floral transition regulator in maize. J. Exp. Bot. 58, 3679–3693. ( 10.1093/jxb/erm217) [DOI] [PubMed] [Google Scholar]
- 50.Haury AC, Mordelet F, Vera-Licona P, Vert JP. 2012. TIGRESS: Trustful Inference of Gene REgulation using Stability Selection. BMC Syst. Biol. 6, 145 ( 10.1186/1752-0509-6-145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altay G, Emmert-Streib F. 2010. Inferring the conservative causal core of gene regulatory networks. BMC Syst. Biol. 4, 132 ( 10.1186/1752-0509-4-132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P. 2010. Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 5, e0012776 ( 10.1371/journal.pone.0012776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yilmaz A, Nishiyama MY Jr, Fuentes BG, Souza GM, Janies D, Gray J, Grotewold E. 2009. GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol. 149, 171–180. ( 10.1104/pp.108.128579) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin J, Tian F, Yang D-C, Meng Y-Q, Kong L, Luo J, Gao G. 2017. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D0140–D1045. ( 10.1093/nar/gkw982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashburner M, et al. et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. ( 10.1038/75556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gene Ontology Consortium 2015. Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–D1056. ( 10.1093/nar/gku1179) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.