Abstract
Dissociating the complexity of metabolic processes into modules is a shift in focus from the single gene/gene product to functional and evolutionary units spanning the scale of biological organization. When viewing the levels of biological organization through this conceptual lens, modules are found across the continuum: domains within proteins, co-regulated groups of functionally associated genes, operons, metabolic pathways and (sub)cellular compartments. Combining modules as components or subsystems of a larger system typically leads to increased complexity and the emergence of new functions. By virtue of their potential for ‘plug and play’ into new contexts, modules can be viewed as units of both evolution and engineering. Through consideration of lessons learned from recent efforts to install new metabolic modules into cells and the emerging understanding of the structure, function and assembly of protein-based organelles, bacterial microcompartments, a structural bioengineering approach is described: one that builds from an architectural vocabulary of protein domains. This bioarchitectonic approach to engineering cellular metabolism can be applied to microbial cell factories, used in the programming of members of synthetic microbial communities or used to attain additional levels of metabolic organization in eukaryotic cells for increasing primary productivity and as the foundation of a green economy.
This article is part of the themed issue ‘Enhancing photosynthesis in crop plants: targets for improvement’.
Keywords: bacterial microcompartment, synthetic biology, carboxysome
1. Modules in biology
The establishment of mitochondria and chloroplasts, dedicated compartments for the reactions of cellular respiration and oxygenic photosynthesis, is a widely regarded key event in the evolution of eukaryotic complexity [1]. These physical structures not only insulate pathways to improve flux, but also serve to confine highly reactive species. Likewise, the evolution of morphological complexity and diversity in higher organisms such as plants and animals has been attributed to hierarchical, modular construction using reiterated parts [2]. This characteristic division of labour in eukaryotic organisms, whether conferred by organelles or from supervening organizational levels of specialized cells, tissues, organs and systems to morphological traits and beyond, can be viewed as nested or hierarchical bioarchitecture; effectively, biological units are combined and organized into progressively more complex, higher order structures. The explanatory power of the concept of recursive modularity has been a major force in unifying the fields of eukaryotic evolution and development into ‘Evo–Devo’ [3–5].
At the same time, it is becoming apparent that the concept of modularity links evolution and bioengineering. Recursive modularity also extends subcellularly and can be used to dissociate the complexity of cellular metabolism into recurring patterns of semi-autonomous functional units: pathways, enzymes, protein domains (figure 1). The nested forms of modularity and hierarchy exhibited by cellular metabolism have been suggested to reflect stages in the emergence of catalytic control by living systems over macromolecular organic chemistry [6]. In contrast with eukaryotic organisms which diversify in cell types and body plans, the evolution of bacteria can be described as a history of metabolic diversification, driven by the relatively facile (compared to endosymbiosis) horizontal transfer of genes and gene clusters. This has a contemporary counterpart in the ability to fabricate DNA and transform and edit genomes, thereby creating new opportunities for engineering modules into cells.
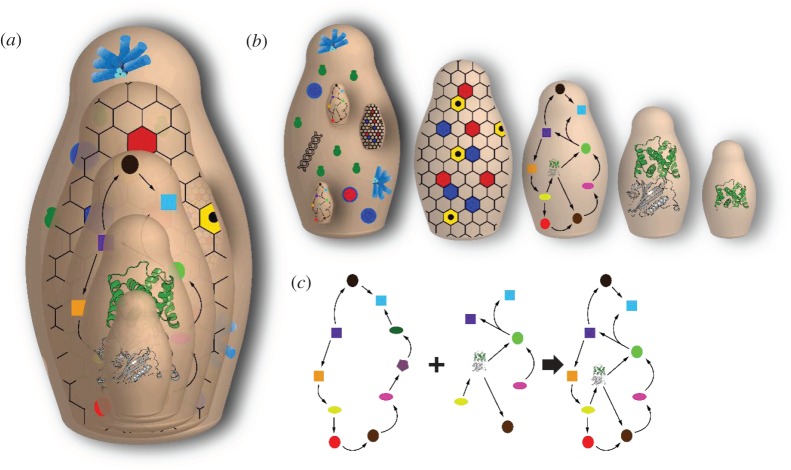
Figure 1.
Russian nesting doll (matryoshka) analogy for recursive modularity in cellular metabolism. (a) The conceptual nesting of modules in a cell such as a cyanobacterium which contains, for example, (b) light harvesting modules, multiple pathways and carboxysomes, which are composed of proteins, which are composed of domains, the basic unit of protein fold and function. (c) Enzyme-centric view of pathways in which enzymes are visualized as nodes and the boundaries of the pathway module are demarcated by the flow of metabolites. Rewiring metabolite flow with the addition of new enzymes and the altering of native connections allows new pathway modules to be constructed. (Online version in colour.)
Understanding the multi-scale modular architecture of cellular metabolism in eukaryotic and prokaryotic cells is a pre-requisite to its engineering. The concept of modularity has been particularly valuable in the merging of engineering and biology into synthetic biology. As a conceptual framework, it simplifies the engineering process by describing functional building blocks according to modular properties. Each module is an end and a means, semi-autonomous, but yet a building block that can be used to increase the complexity of function and adaptation to new contexts and environments. In synthetic biology, molecular elements can be categorized as ‘parts' (the lowest level of the hierarchy) that can be used to construct devices (parts assembled together to yield a desired function), which can in turn be further combined into devices or modules.
Implicit in modular bioengineering is the idea that biological systems are composed of functional modules that can be individually swapped or inserted into other systems, leading to a change in the overall characteristics of the system. This ability to affect the system is predicated on the ability to integrate the new module with the extant metabolism. As in the ancient symbiosis leading to the chloroplast, connecting the module to the host system to enable its function is critical. At the same time, the module will fail if it causes too extreme perturbation of the native metabolism; the means for interfacing of a newly introduced module with the host metabolism is a key design requirement, comparable in importance to the design of the module itself.
Recently, there have been several advances in engineering bacterial cells with metabolic modules in the form of pathways. The studies referred to here focus on improving CO2 fixation (only a few examples are chosen; this is not intended to be a review of the literature). They underscore the challenges associated with introducing a module that must simultaneously interface with, and be insulated from, portions of the host metabolism. Physical compartmentalization, as in eukaryotic organelles, is useful to gain control over this balance.
Unlike eukaryotic organelles, which rely on lipid-based membranes, bacterial microcompartments (BMCs) such as carboxysomes offer a design template for building multi-enzyme-containing organelles that can be encoded by a DNA blueprint [7]. The selectively permeable ‘membranes’ of BMCs are composed of proteins that self-assemble with the constituent enzymes. Recent progress in the structural biology of BMCs has provided the requisite physical description of the building blocks, which, combined with a knowledge of their functions and their interactions in BMC assembly, makes the goal of designing and building dedicated protein-based compartments attainable [7–10]. Such a bioarchitectonic approach coupled with advances in DNA fabrication, genome editing and ancestral sequence reconstruction [11] to resurrect relatively promiscious enzymatic activities [12,13], coupled with laboratory evolution, provides unprecedented opportunities to endow cells with new modules to increase their complexity, and concomitantly their programmable control.
2. Modular engineering of metabolic pathways and cycles
Although the organelles of eukaryotic cells as structurally defined metabolic spaces are arguably the most recognizable metabolic modules, a subcellular module does not have to be a single physical entity defined in time and space but may be conceptualized as a pathway or a process [14]. This is apparent if one visualizes pathways with an enzyme-centric view, rendering the enzymes as the nodes connected by the successive substrates and products (figure 1c). From this perspective, the movement of metabolites within the pathway abstractly demarcates the module boundaries. At the same time, pathway modules are linked to one another by metabolites, invoking a basic requirement of modular engineering: design and construction of semi-autonomous functional units must include the means for inter-module connections.
Installed, heterologous pathways need the requisite substrates for their function, typically by draining metabolites from native host pathways. But with the increased demand for metabolites in an engineered cell, the supply may be insufficient, causing the perturbation of the function of native pathways, or simply resulting in too little input for the installed pathway to function. This was evident in our recent attempt to improve photorespiration in cyanobacteria [15]. Given the intractability of improving Rubisco, the underlying motivation was to build a synthetic pathway with several advantages over naturally occurring salvage pathways. In comparison with natural photorespiratory pathways, the synthetic bypass would not only prevent the loss of NH3 but also result in a net gain in carbon fixation rather than a net loss. The designed pathway used oxygen-tolerant enzymes that are components of the 3-hydroxypropionate bi-cycle, an alternative autotrophic CO2 fixation pathway found in the green, non-sulfur thermophile Chloroflexus aurantiacus [16]. One turn of the designed cyclic photorespiratory bypass consumes glyoxylate, fixes bicarbonate and ultimately leads to the production of pyruvate which can be used directly for biosynthesis or to replenish the Calvin–Benson–Bassham (CBB) cycle.
To build the synthetic salvage module to reassimilate the photorespiratory by-product glyoxylate, six genes, encoded in assembled DNA constructs spanning more than 16 kb, were required [15]. All of the enzymes were shown to be active in cyanobacterial extracts. However, there was no resulting phenotype, even under conditions known to exacerbate photorespiration. The bottleneck was traced by examining reactions rates of enzymes that produce metabolites to feed the installed module [15]. For example, the measured activity level for native acetyl-CoA carboxylase, which feeds substrate into the synthetic bypass, was close to or below the detection limit of the assay, suggesting that manipulating this enzyme to increase its product levels would be needed for it to support both native metabolism and the installed module.
In addition to engineering photorespiration, other efforts to enhance CO2 fixation have sought to circumvent the involvement of Rubisco altogether. Various synthetic carbon fixation pathways have been designed computationally by mining parts from enzyme databases and building pathways in silico [17]. Another recently reported approach to engineering autotrophy involved repurposing enzymes already available in the host organism [18]. Using computational approaches, Antonovsky et al. inventoried Escherichia coli for ‘parts’ that could be used to construct the CBB cycle for CO2 fixation. Indeed, most of the reactions of the CBB cycle take place in the context of glycolysis and the pentose phosphate pathway, albeit in the reverse direction relative to their role in the CBB cycle. Only the carboxylation of RuBP by Rubisco to form 3-phosphoglycerate is unique to the CBB cycle. In the design, the enzymes of gluconeogenesis were dissociated from their native pathways by severing connections among the native E. coli pathways (metabolic modules). The disconnected enzymes could then be constellated into a new functional module (figure 1c), with the addition of the enzymes not native to E. coli (phosphoribulokinase and Rubisco). However, similar to the attempt to install the synthetic photorespiratory bypass, the CO2 fixation module was tolerated, but not able to render the E. coli capable of growth on CO2 alone. Laboratory evolution was subsequently imposed to force the engineered E. coli to rely on the CO2 fixation module. Interestingly, by sequencing the genomes of the evolved strains, it was found that the mutations were in genes for enzymes that controlled flux points from the module, not in the parts of the module itself. The results underscore the importance of metabolites in ‘structuring’ module boundaries and inter-module connections; unwanted cross-talk accounts for a significant challenge in the design and construction of synthetic pathway modules.
3. Physically insulated metabolic modules in bacteria: bacterial microcompartments
As these examples illustrate, metabolites must achieve an exquisite balance between demarcating module boundaries, and providing inter-module communication. Structuring and enclosing metabolic space with a selectively permeable membrane offers a solution for achieving this balance. In organelles, a physical barrier such as a lipid bilayer establishes the module boundary and by tuning the permeability of this barrier metabolites can then be selectively partitioned into a pool confined to the module and those that establish connections among modules in the cytosol.
The presence of organelles has long been considered a key distinction between prokaryotic and eukaryotic cells. However, already more than 50 years ago, there were hints of higher order organization in bacterial metabolism with the discovery, by electron microscopy, of carboxysomes in cyanobacteria [19] (figure 2). These large (approx. 100–600 nm diameter [21]) polyhedral bodies sequester carbonic anhydrase and Rubisco within a protein shell, thereby concentrating substrates and protecting Rubisco from oxygen generated by the light reactions (figure 3a). Two distinct types of carboxysomes are known, α and β, encapsulating Form 1A and Form 1B Rubisco, respectively [22].
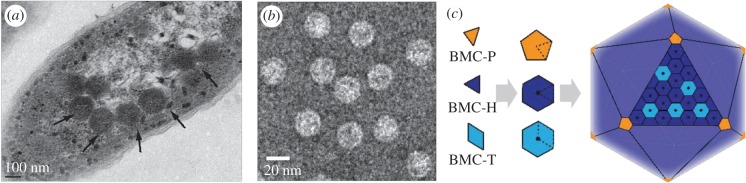
Figure 2.
BMC architectures. (a) Electron micrograph of the cyanobacterium Synechococcus PCC7942 containing numerous carboxysomes (arrows). (b) Empty β carboxysome shells produced in and purified from E. coli, stained with ammonium molybdate and visualized on an electron microscopy grid (see [20] for details). (c) Schematic of the structural elements of bacterial microcompartment shells. Electron micrographs courtesy of Dr Fei Cai. (Online version in colour.)
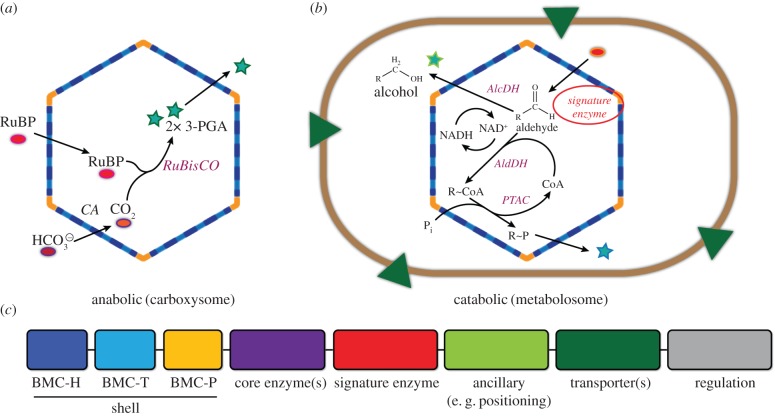
Figure 3.
BMCs are metabolic modules encoded by genetic modules. Schematics of (a) carboxysome and (b) metabolosome function. (c) Genes for the structural components of the BMC well as gene products providing supporting functions (e.g. transporters for bringing substrates into the cell; cytoskeletal elements for positioning the organelle) are typically encoded together in a genetic module. (Online version in colour.)
The carboxysome was long thought to be a peculiarity confined to cyanobacteria. It is now known, by the detection of homologues to carboxysome shell protein genes in sequenced bacterial genomes, that such BMCs are widespread among the bacteria. Common to all BMCs is a shell composed of homologous proteins that form hexamers (BMC-H proteins), trimers that form pseudohexamers (BMC-T proteins) and pentamers (BMC-P proteins) (figure 2c). A comprehensive identification and taxonomy of BMCs, based on their component parts which were defined as protein domains, was compiled in 2014, revealing an array of different functional types, found across 23 bacterial phyla [23]. With the increasing emphasis on sequencing of diverse microbial genomes, new candidate phyla and microbial dark matter have undoubtedly added to the number and the diversity of BMCs to be found in databases.
BMCs colocalize enzymes constituting segments of metabolic pathways to enhance flux (figure 3). The majority of BMCs are not involved in CO2 fixation but are involved in catabolic reactions. Many of the latter, also referred to as metabolosomes, share a metabolic core (figure 3b) composed of an acylating aldehyde dehydrogenase, an alcohol dehydrogenase and a phosphotransacylase [7,23]. These three enzymes work in concert to metabolize a substrate generated by a signature enzyme, for example, ethanolamine ammonia lyase or propanediol dehydratase. BMCs share the hallmark attributes of membrane-bound organelles, confining toxic metabolites, functioning to insulate reactions from competing pathways, and protecting labile enzymes, cofactors and damage-prone metabolites by establishing a controlled environment within a selectively permeable protein shell. However, there is a fundamental difference between the permeability properties of membrane-bound organelles and the shells of BMCs; they have inverse selectivity. BMCs are permeable to small charged and polar substrates (the pores in shell proteins range from 4 to 14 Å [24,25]) and are relatively impermeable to non-polar molecules, such as CO2 [26,27].
BMCs are typically encoded in loci that contain genes not only for the shell and the component enzymes, but also for membrane proteins presumed to be important for cellular uptake of the substrate(s) for the organelle. Putative cytoskeletal elements for positioning the organelle (figure 3c) are frequently also found in these loci, suggesting that, as for chloroplasts, the subcellular location/distribution of BMCs is important to function. The obvious rampant horizontal gene transfer of BMC loci [23] may be reflective of the adaptive value of compactly organizing the structural and ancillary genes for BMC function, enabling their incorporation into host metabolism. Indeed, prokaryotic gene clusters such as those encoding BMCs have been proposed to constitute ready-made genetic modules for plug and play applications in synthetic biology [28]. The carboxysome, for example, has attracted extensive attention for its potential to confer or enhance the capacity for CO2 fixation by Rubisco. Early efforts by researchers to transfer BMCs across species focused on structural components of the organelles, achieving some success. The core locus of the α carboxysome has been expressed in E. coli and the associated Rubisco was shown to be active [29]; likewise the propanediol utilization microcompartment was fully functional when genes for its assembly were transferred from the native host (Citrobacter) to E. coli [30]. In the first demonstration of heterologous expression of BMC proteins across kingdoms, shell protein genes as well as core components of the β carboxysome, including cyanobacterial Rubisco, have been expressed in chloroplasts [31,32].
4. Assembly and engineering of the carboxysome
In addition to using naturally occurring BMCs such as carboxysomes for metabolic engineering, BMCs provide a design template for building protein-based organelles that could be used to insulate heterologous pathways to prevent cross-talk, to establish private cofactor pools or to spatially colocalize enzymes to improve flux [7–9]. Understanding how BMCs self-assemble is fundamental to efforts to take advantage of this architecture in bioengineering. Do they assemble like eukaryotic organelles, in which component proteins are targeted to a pre-formed compartment? Or does the core assemble first, followed by encapsulation within a shell? Interestingly, there appear to be two different modes of BMC biogenesis, one involving co-assembly of shell and core enzymes, exemplified by the α carboxysome [33,34]. The second type, demonstrated for the β carboxysome, in which the catalytic core assembles first followed by encapsulation, is proposed to also apply to the majority of metabolosomes [7,35–37].
Despite the importance of the core enzymes for either mode of assembly, empty BMC shells can self-assemble when the constituent proteins are expressed in E. coli [20,38,39]. These tend to be smaller than their packed counterparts (figure 2b). Such empty shells could serve as three-dimensional, polyvalent scaffolds for enzymes, achieved by fusion of enzymes to the constituent shell proteins. This will perhaps require a mixture of native BMC-H proteins as well as their fusion counterparts; this was, for example, necessary when shell proteins were modified with GFP [35]. It seems likely that this is required because steric hindrance by the fluorophore precludes formation of a hexamer or the assembly of hexamers into a facet.
While it is possible to target proteins to BMC shells using encapsulation peptides which are thought to mediate interaction between the enzymatic cargo and the shell [37,40,41], designs that compartmentalize multiple enzymes take full advantage of the potential of BMCs. As proof-of-principle of the potential for designing BMC catalytic cores based on knowledge of the structure and interactions of protein domains, we undertook a re-design to simplify the β carboxysome core; the re-engineering also provided a test of our understanding of the principles of assembly for the β carboxysome. Using knowledge of the sequence of protein domain interactions in the course of carboxysome biogenesis, a synthetic gene for a chimeric protein was designed that functionally replaces four different gene products in carboxysome assembly [42]. Only a subset of the native complement of protein domains in the carboxysome core was selected, and their coding sequences fused together in a specific order anticipated to preserve critical interactions in carboxysome assembly. The resulting chimeric protein combined domains for Rubisco nucleation, and carbonic anhydrase activity followed by an encapsulation peptide for interaction with the shell [36,37]. Notably, the design of the synthetic core protein rendered some of the native protein domains dispensable, thereby reducing the amount of DNA needed and reducing the number of genes necessary to form a carboxysome core with Rubisco from four to one. The resulting streamlined carboxysomes were shown to support photosynthesis in cyanobacteria. Because there is a clear read-out for both structure (visible in micrographs) and function (photoautotrophic growth in air, without additional CO2 supplementation), the β carboxysome provided an ideal model system for demonstrating the potential for domain-based re-engineering of BMC cores.
5. Bacterial microcompartment architectonics: engineering metabolic modules based on domain folds and functions
Several lines of evidence (summarized in [7]) suggest that the majority of BMCs assemble as a series of interacting protein domains that coalesce and are subsequently surrounded by the shell. Given that protein domains are the structural, functional and evolutionary units of proteins, (re-)engineering BMCs and other large multi-protein macromolecular assemblies may be tractable by focusing on domain structures and interactions, leveraging the inherent modularity of proteins for building new subcellular architectures. In this approach, protein domains constitute the architectural vocabulary for constructing new kinds of metabolic compartments. In practice, such a structure-based bioarchitectonic strategy involves surveying encapsulated protein folds in known BMCs to serve as the set of building blocks for engineering catalytic activities. In general, the functions supported by fold families are typically broad; diverse catalytic activities are frequently supported by the same structural scaffold [43–45]. The TIM barrel fold found in several types of BMCs [23], for example, carries out hundreds of different kinds of catalytic reactions in nature [43]. The diverse protein fold families that assemble into BMCs can accordingly be viewed as a set of generic building blocks that can be customized for function.
A fold-based construction of new kinds of BMC cores using naturally encapsulated folds can be approached in two distinct ways; either conversion of a natively encapsulated enzyme into a member of the same fold family with the desired activity (i.e. active site conversion), or, by contrast, taking a cytosolic enzyme and endowing it with the attributes of an encapsulated member of the same superfamily. For example, several different types of BMCs encapsulate glycyl radical enzymes (GREs) with diverse functions [46]. Bioinformatic analysis in conjunction with structural modelling could be used to identify features of an encapsulated B12-independent diol dehydratase [47] (a member of the GRE family of enzymes) likely to be important for encapsulation by comparison with its non-BMC-associated counterparts. The identified encapsulation peptides and surface features for domain interactions could be engineered into pyruvate formate lyase (PFL), a cytosolic enzyme that is a member of the GRE family, to endow it with the necessary structural attributes (e.g. an encapsulation peptide) to promote packaging into a BMC. This could be useful, for example, in promoting formatrophic growth of E. coli [48,49].
A second strategy involves converting the active site of a naturally encapsulated GRE into that of a PFL. While making systematic point mutations to convert the active site of an encapsulated GRE into that of PFL sounds simple, in practice such active site transformations have proven challenging, with the reasons for failure often not readily apparent. An alternative approach would be to use ancestral sequence reconstruction methods to resurrect a primordial encapsulated GRE sequence followed by laboratory evolution to canalize the specificity of its reaction [44]. The cyanobacterial carboxysome, with its sensitive read-out for form and function, again provides an ideal model system to test this general strategy. A recent report described how resurrected, ancestral Rubisco large subunits could be packaged into cyanobacterial carboxysome shells [50], demonstrating how ancestral sequence reconstruction can potentially be combined with BMC engineering.
In conjunction with constructing catalytic cores from protein domains, designing physically compartmentalized metabolic modules also requires tailoring the permeability properties of the protein shell to support the encapsulated reactions. Shell protein hexamers and pseudohexamers are typically perforated by pores that allow diffusion of metabolites into the BMC (figure 2c). Several studies have probed the underlying determinants of this selectivity [51,52]. Manipulation of both the size and the charge of the residues surrounding the pores can be used to gain control over their selective permeability. Recently, advances have been made in endowing the shell with a function beyond serving as a selectively permeable barrier. In a step towards building synthetic BMCs for redox reactions, shell proteins have been engineered to bind iron–sulfur clusters [53]. The conversion of a passive barrier into a redox active membrane represents a major advance in the construction of tailor-made nanoreactors for biotechnological applications that can be connected via electron reactions with the rest of cellular metabolism.
The ability to transfer electrons and other high-energy intermediates between BMCs and the cytosol expands the range of nanoreactors that could conceivably be designed. Would it be possible to build a BMC for nitrogen fixation—a nitrogenosome? Given the observation that ATP-dependent enzymes are associated with BMCs [7,46,54], it seems that ATP can cross the shell in some as yet uncharacterized way. For a nitrogenosome core, the carboxysome assembly pathway provides a model for how nitrogenase could be assembled first, followed by encapsulation within a shell. Cofactors would be brought into the lumen via their cognate proteins, as was recently shown for the metabolosome-associated phosphotransacylase [55]. Domain fusions could be used to tether proteins together, as demonstrated for the chimeric carboxysome core protein [42]. Nitrogenase presents an interesting additional design challenge, the need to accommodate large domain motions during catalysis; this could be addressed by domain fission, separating the domains of nitrogenase into discrete proteins that would, nevertheless, remain in close spatial proximity due to confinement within the shell. Recently, it was shown that it is possible to split the domains of an encapsulated GRE into separate polypeptides and still retain full catalytic activity [47]. In all of these examples, protein domains provide the structural and functional foundation of a bioarchitectonic approach to engineering BMCs.
6. Conclusion and prospects
Looking forward, engineering designed metabolic modules could be used to metabolically programme members of synthetic microbial consortia. Such modules could be used to help divide the ‘labour’ among the members of a designed multi-cellular community composed of different types of bacteria [56]. Likewise, in the context of microbial cell factories, synthetic BMCs could be used to insulate non-native pathways, including those constructed from partitioned building blocks of native metabolism, as in the hemi-autotrophic E. coli [18]. For eukaryotic organisms, in addition to efforts focused on installing carboxysomes in chloroplasts [31,32], compartmentalization of other pathways within mitochondria or chloroplasts could provide protection from local side reactions caused by highly reactive intermediates, which cause enzymatic mistakes that generate wasteful and toxic side-products [57]. Given that the permeability of the protein shell is the inverse of that of a lipid membrane, encapsulation in BMC shells could be used to further organize reactions within eukaryotic organelles to prevent cross-talk among native and introduced pathways. The ability to build compartments within compartments such as the chloroplast provides an opportunity to further refine the hierarchical construction of metabolism. Such engineered bioarchitecture, in which modules are both ends and means, recapitulates a hallmark attribute of biological systems. Indeed, conceptually, it underlies the genesis of the terms organism and organization in the context of living systems [58]. Modular engineering of biology provides an opportunity to gain a new degree of control over metabolism to address grand challenges of our time, such as food and fuel security, and provide the foundation for a bioeconomy.
Acknowledgements
I thank Dr Fei Cai and Dr Markus Sutter for assistance with the figures and critical readings of the manuscript.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID) grant 1R01AI114975-01 and the US Department of Energy, Basic Energy Sciences, Contract DE-FG02-91ER20021 with infrastructure support from MSU AgBio Research.
References
- 1.Archibald JM. 2015. Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 25, R911–R921. ( 10.1016/j.cub.2015.07.055) [DOI] [PubMed] [Google Scholar]
- 2.Carroll S. 2001. Chance and necessity: the evolution of morphological complexity and diversity. Nature 409, 1102–1109. ( 10.1038/35059227) [DOI] [PubMed] [Google Scholar]
- 3.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. 1999. From molecular to modular cell biology. Nature 402, C47–C51. ( 10.1038/35011540) [DOI] [PubMed] [Google Scholar]
- 4.Winther R. 2001. Varieties of modules: kinds, levels, origins, and behaviors. J. Exp. Zool. 291, 116–129. ( 10.1002/jez.1064) [DOI] [PubMed] [Google Scholar]
- 5.Sherker JM, Lucks JB, Arkin AP. 2009. Evolution, ecology and the engineered organism: lessons for synthetic biology. Genome Biol. 209, 114 ( 10.1186/gb-2009-10-11-114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braakman R, Smith E. 2013. The compositional and evolutionary logic of metabolism. Phys. Biol. 10, 011001 ( 10.1088/1478-3975/10/1/011001) [DOI] [PubMed] [Google Scholar]
- 7.Kerfeld CA, Erbilgin O. 2015. Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol. 23, 22–34. ( 10.1016/j.tim.2014.10.003) [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Esquer CR, Newnham SE, Kerfeld CA. 2016. Bacterial microcompartments as metabolic modules for plant synthetic biology. Plant J. 87, 66–75. ( 10.1111/tpj.13166) [DOI] [PubMed] [Google Scholar]
- 9.Frank SLA, Prentice MB, Warren MJ. 2013. Bacterial microcompartments moving into a synthetic biological world. J. Biotechnol. 163, 273–279. ( 10.1016/j.jbiotec.2012.09.002) [DOI] [PubMed] [Google Scholar]
- 10.Wang PLE, Zhao FJ, Kopittke PM. 2016. Nanotechnology: a new opportunity in plant sciences. Trends Plant Sci. 21, 699–712. ( 10.1016/j.tplants.2016.04.005) [DOI] [PubMed] [Google Scholar]
- 11.Gumulya Y, Gillam EMJ. 2017. Exploring the past and the future of protein evolution with ancestral sequence reconstruction: the ‘retro’ approach to protein engineering. Biochem. J. 474, 1–19. ( 10.1042/BCJ20160507) [DOI] [PubMed] [Google Scholar]
- 12.Tawfik OKDS. 2010. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505. ( 10.1146/annurev-biochem-030409-143718) [DOI] [PubMed] [Google Scholar]
- 13.Copley SD. 2015. An evolutionary biochemist's perspective on promiscuity. Trends Biochem. Sci. 40, 72–78. ( 10.1016/j.tibs.2014.12.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira-Leal JB, Levy ED, Teichmann SA. 2006. The origins and evolution of functional modules: lessons from protein complexes. Phil. Trans. R. Soc. B 361, 507–517. ( 10.1098/rstb.2005.1807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih PM, Zarzycki J, Niyogi KK. 2014. Introduction of a synthetic CO2-fixing photorespiratory bypass into a cyanobacterium. J. Biol. Chem. 289, 9493–9500. ( 10.1074/jbc.C113.543132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zarzycki J, Brecht V, Müller M, Fuchs G. 2009. Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proc. Natl Acad. Sci. USA 106, 21 317–21 322. ( 10.1073/pnas.0908356106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bar-Even A, Noor E, Lewis NE, Milo R. 2010. Design and analysis of synthetic carbon fixation pathways. Proc. Natl Acad. Sci. USA 107, 8889–8894. ( 10.1073/pnas.0907176107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonovsky N, et al. 2016. Sugar synthesis from CO2 in Escherichia coli. Cell 166, 115–125. ( 10.1016/j.cell.2016.05.064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drews G, Niklowitz W. 1956. Cytology of Cyanophycea. II. Centroplasm and granular inclusions of Phormidium uncinatum. Arch. Mikrobiol. 24, 147–162. ( 10.1007/BF00408629) [DOI] [PubMed] [Google Scholar]
- 20.Cai F, Bernstein SL, Wilson SC, Kerfeld CA. 2016. Production and characterization of synthetic carboxysome shells with incorporated luminal proteins. Plant Physiol. 170, 1868–1877. ( 10.1104/pp.15.01822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Esquer CR, Smarda J, Rippka R, Axen SD, Guglielmi G, Gugger M, Kerfeld CA. 2016. Cyanobacterial ultrastructure in light of genomic sequence data. Photosynth. Res. 29, 147–157. ( 10.1007/s11120-016-0286-2) [DOI] [PubMed] [Google Scholar]
- 22.Kerfeld CA, Melnicki MR. 2016. Assembly, function and evolution of cyanobacterial carboxysomes. Curr. Opin. Plant Biol. 31, 66–75. ( 10.1016/j.pbi.2016.03.009) [DOI] [PubMed] [Google Scholar]
- 23.Axen SD, Erbilgin O, Kerfeld CA. 2014. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput. Biol. 10, e1003898 ( 10.1371/journal.pcbi.1003898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. 2009. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 392, 319–333. ( 10.1016/j.jmb.2009.03.056) [DOI] [PubMed] [Google Scholar]
- 25.Kerfeld CA, et al. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309, 936–938. ( 10.1126/science.1113397) [DOI] [PubMed] [Google Scholar]
- 26.Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S. 2009. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS ONE 4, e7521 ( 10.1371/journal.pone.0007521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, Cannon GC. 2008. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Biol. Chem. 283, 10 377–10 384. ( 10.1074/jbc.M709285200) [DOI] [PubMed] [Google Scholar]
- 28.Fischbach M, Voight C. 2010. Prokaryotic gene clusters: a rich toolbox for synthetic biology. Biotechnol. J. 5, 1277–1296. ( 10.1002/biot.201000181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonacci WTP, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF. 2012. Modularity of a carbon-fixing protein organelle. Proc. Natl Acad. Sci. USA 109, 478–483. ( 10.1073/pnas.1108557109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons JBDS, et al. 2008. Biochemical and structural insights into bacterial organelle form and biogenesis. J. Biol. Chem. 283, 14 366–14 375. ( 10.1074/jbc.M709214200) [DOI] [PubMed] [Google Scholar]
- 31.Lin OA, Andralojc PJ, Devonshire J, Hines KM, Parry MA, Hanson MR. 2014. β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 79, 1–12. ( 10.1111/tpj.12536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin OA, Andralojc PJ, Parry MA, Hanson MR. 2014. A faster Rubisco with potential to increase photosynthesis in crops. Nature 513, 547–550. ( 10.1038/nature13776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A. 2007. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J. Mol. Biol. 372, 764–773. ( 10.1016/j.jmb.2007.06.059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ. 2010. Organization, structure, and assembly of α-carboxysomes determined by electron cryotomography of intact cells. J. Mol. Biol. 396, 105–117. ( 10.1016/j.jmb.2009.11.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. 2013. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155, 1131–1140. ( 10.1016/j.cell.2013.10.044) [DOI] [PubMed] [Google Scholar]
- 36.Aussignargues C, Paasch BC, Gonzalez-Esquer R, Erbilgin O, Kerfeld CA. 2015. Bacterial microcompartment assembly: the key role of encapsulation peptides. Commun. Integr. Biol. 8, e1039755 ( 10.1080/19420889.2015.1039755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinney JN, Salmeen A, Cai F, Kerfeld CA. 2012. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J. Biol. Chem. 287, 17 729–17 736. ( 10.1074/jbc.M112.355305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons JBFS, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ. 2010. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol. Cell. 38, 305–315. ( 10.1016/j.molcel.2010.04.008) [DOI] [PubMed] [Google Scholar]
- 39.Lassila JK, Bernstein SL, Axen SD, Kinney JN, Kerfeld CA. 2014. Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. J. Mol. Biol. 426, 2217–2228. ( 10.1016/j.jmb.2014.02.025) [DOI] [PubMed] [Google Scholar]
- 40.Lawrence AD, et al. 2014. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth. Biol. 3, 454–465. ( 10.1021/sb4001118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan CCS, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. 2010. Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl Acad. Sci. USA 107, 7509–7514. ( 10.1073/pnas.0913199107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez-Esquer CR, Shubitowski TB, Kerfeld CA. 2015. Streamlined construction of the cyanobacterial CO2-fixing organelle via protein domain fusions for use in plant synthetic biology. Plant Cell 27, 2637–2644. ( 10.1105/tpc.15.00329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toth-Petroczy A, Tawfik DS. 2014. The robustness and innovability of protein folds. Curr. Opin. Struct. Biol. 26, 131–138. ( 10.1016/j.sbi.2014.06.007) [DOI] [PubMed] [Google Scholar]
- 44.Bloom JD, Arnold FH. 2009. In the light of directed evolution: pathways of adaptive protein evolution. Proc. Natl Acad. Sci. USA 106(Suppl. 1), 9995–10 000. ( 10.1073/pnas.0901522106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandya C, Farelli JD, Dunaway-Mariano D, Allen KN. 2014. Enzyme promiscuity: engine of evolutionary innovation. J. Biol. Chem. 289, 30 229–30 236. ( 10.1074/jbc.R114.572990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zarzycki J, Erbilgin O, Kerfeld CA. 2015. Bioinformatic characterization of a major family of bacterial organelles: glycyl radical enzyme-associated bacterial microcompartments. Appl. Environ. Microbiol. 81, 8315–8329. ( 10.1128/AEM.02587-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarzycki J, Sutter M, Socorro N, Cortina DJ, Erb TJ, Kerfeld CA. 2017. In vitro characterization and concerted function of three core enzymes of a glycyl radical enzyme—associated bacterial microcompartment. Sci. Rep. 7, 42757 ( 10.1038/srep42757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bar-Even A, Noor E, Flamholz A, Milo R. 2013. Design and analysis of metabolic pathways supporting formatotrophic growth for electricity-dependent cultivation of microbes. Biochim. Biophys. Acta 1827, 1039–1047. ( 10.1016/j.bbabio.2012.10.013) [DOI] [PubMed] [Google Scholar]
- 49.Zelcbuch L, Lindner SN, Zegman Y, Vainberg Slutskin I, Antonovsky N, Gleizer S, Milo R, Bar-Even A. 2016. Pyruvate formate-lyase enables efficient growth of Escherichia coli on acetate and formate. Biochemistry 55, 3851–3863. ( 10.1021/acs.biochem.6b00184) [DOI] [PubMed] [Google Scholar]
- 50.Shih PM, Occhialini A, Cameron JC, Andralojc PJ, Parry MA, Kerfeld CA. 2016. Biochemical characterization of predicted Precambrian RuBisCO. Nat. Commun. 7, 10382 ( 10.1038/ncomms10382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chowdhury CCS, Pang A, Sawaya MR, Sinha S, Yeates TO, Bobik TA. 2015. Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proc. Natl Acad. Sci. USA 112, 2990–2995. ( 10.1073/pnas.1423672112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA. 2015. Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells. ACS Synth. Biol. 4, 444–453. ( 10.1021/sb500226j) [DOI] [PubMed] [Google Scholar]
- 53.Aussignargues CPM, et al. 2016. Structure and function of a bacterial microcompartment shell protein engineered to bind a [4Fe-4S] cluster. J. Am. Chem. Soc. 138, 5262–5270. ( 10.1021/jacs.5b11734) [DOI] [PubMed] [Google Scholar]
- 54.Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. 2013. Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 64, 787–798. ( 10.1093/jxb/ers294) [DOI] [PubMed] [Google Scholar]
- 55.Erbilgin O, Sutter M, Kerfeld CA. 2016. The structural basis of coenzyme A recycling in a bacterial organelle. PLoS Biol. 14, e1002399 ( 10.1371/journal.pbio.1002399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou K, Qiao K, Edgar S, Stephanopoulos G. 2015. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 33, 377–383. ( 10.1038/nbt.3095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanson AD, Henry CS, Fiehn O, Lagard V. 2016. Metabolite damage and metabolite damage control in plants. Annu. Rev. Plant Biol. 67, 131–152. ( 10.1146/annurev-arplant-043015-111648) [DOI] [PubMed] [Google Scholar]
- 58.Fox KE. 2007. The disappearance of function from ‘self-organizing systems'. In Systems biology (eds Boogerd F, Bruggeman F, Hofmeyer J, Westerhoiff H), pp. 303–317. Amsterdam, The Netherlands: Elsevier BV. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.