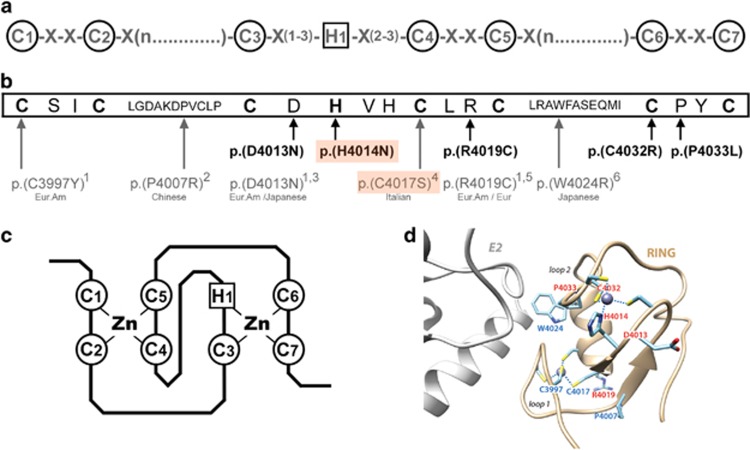
Figure 3.
Rare RNF213 variants of MMA patients located in the RING-finger domain. (a) C3HC4 RING-finger consensus motif.27 Highly conserved cysteine and histidine residues involved in zinc-coordination are circled. (b) RNF213 RING-finger sequence (NP_001243000.2). Highly conserved cysteine and histidine residues are in bold. The rare RNF213 variants identified in MMA patients and involving the RING domain are shown below the RING-finger sequence. Variants found in our cohort of 68 Caucasian MMA are represented in bold. Those found in previously published MMA cohorts are represented in gray, and ethnicities of the corresponding cases are given below the variants. (1: Cecchi et al, Stroke, 2014; 2: Wu et al. PLoS One, 2012; 3: Liu et al. PLoS One, 2011; 4: Raso et al. J Neurosurg Sci, 2016; 5: Kobayashi et al. PLoS One, 2016; 6: Miyatake et al. Neurology, 2012). The p.(H4014N) and p.(C4017S) de novo variants are boxed in orange. Eur, European; Eur. Am, European American (c) Representation of the ‘cross-brace’ structure of the C3HC4 RING-finger domain, mediated through cysteine and histidine Zinc-binding. Panel D: Model of the 3D structure of the RNF213 RING finger (ribbon representation), based on the 3D structure of the RING domain of RNF146 (pdb 4qpl), solved in complex with its E2.43 Amino acids which are mutated in moyamoya patients are labeled in red (our study) and blue (previous studies). p.(R4019C) and p.(D4013N), found mutated both in our study and previous studies, are in red. The position of the putative E2 partner is shown in gray, in reference to the structure of the E2-E3 complex reported in DaRosa et al.,43 with which the model of the RN213 RING 3D structure was superimposed.