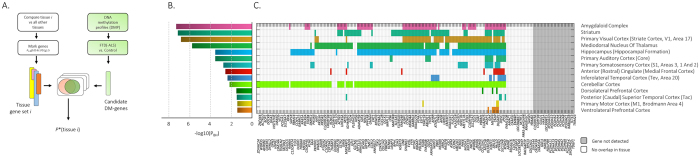
Figure 2.
Tissue enrichment for FTD-ALS based on the brain tissue specific DNA-methylation profiles of BrainSpan. (A) The hypergeometric test is used to compute P-value for each tissue based on the following parameters; total number of genes (M), number of tissue specific genes (K), number of significant differential methylated genes (N), and the overlap of significant differential methylated genes and the genes in the tissue specific gene set (x). The final P-value (P*) is corrected for multiple testing using Benjamini and Hochberg, and used for tissue selection with PBH < 0.05. (B) Enriched tissues in FTD-ALS sorted in -log10(PBH). (C) Genes are colored with the brain tissue specific color if overlap is seen with any of the significantly differential DNA-methylated genes in FTD-ALS.