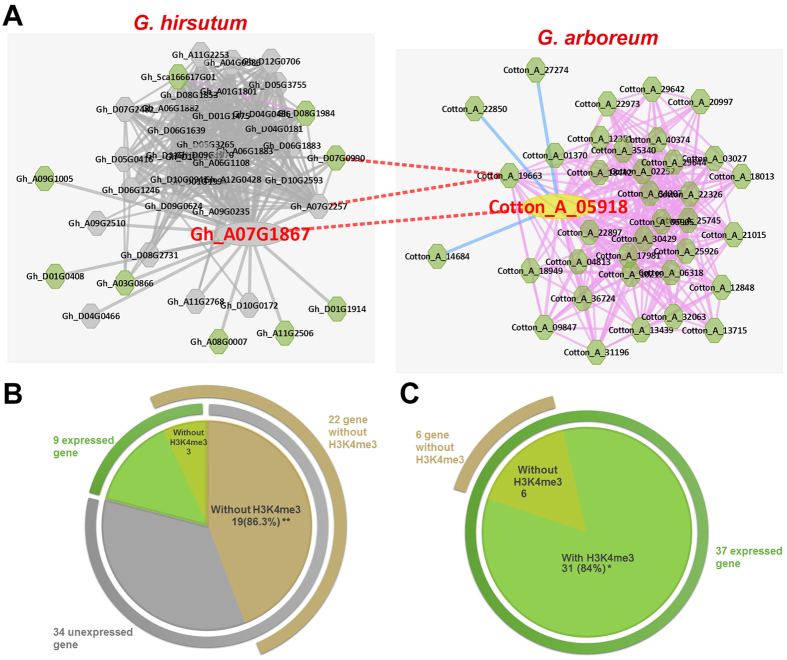
Figure 9.
Modularized comparison of differential histone modification during evolution. (A) The co-expression networks of the orthologous gene pair of Gh_A07G1867 and Cotton_A_05918 are compared. Grey and green nodes are unexpressed and expressed genes in root tissues, respectively. A pink or blue line links positive co-expressed or negative co-expressed gene pairs, respectively. A red dotted line links orthologous gene pairs. (B) There are 43 co-expressed genes in the root-preferential co-expression network of Gh_A07G1867, including 9 expressed genes (green loop covers) and 34 unexpressed genes (grey loop covers). Among them, 22 co-expressed genes lack the H3K4me3 histone modification. In G. hirsutum, 50,113 genes had the H3K4me3 modification and 20,365 genes lacked it. In the sub-network, 34 genes were unexpressed in root, and among them, 15 genes had the H3K4me3 and 19 genes did not. Fisher’s Exact Test was performed, and p-values of 0.001 are marked by **. (C) There are 37 co-expressed genes in the root-preferential co-expression network of Cotton_A_05918, and all of them are expressed in root. Among these, 6 co-expressed genes did not have H3K4me3 histone modification. In G. arboreum, 28,923 genes had the H3K4me3 modification and 12,408 genes did not. In the sub-network, 37 genes were expressed in root and among them, 31 genes had the H3K4me3 and 6 genes did not. Fisher’s Exact Test was performed, and the p-value was 0.073 and marked by *.