Figure 2.
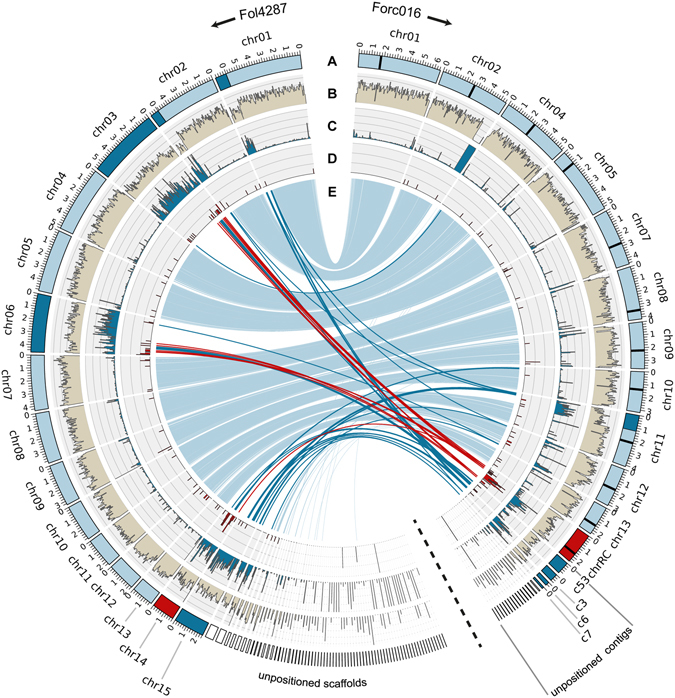
The SMRT genome assembly of Forc016 includes eleven core chromosomes, several repeat-rich, gene-poor accessory regions and one chromosome enriched in candidate effector genes. Comparison of the Forc016 assembly to that of Fol4287 reveals (A) eleven conserved core chromosomes (light blue), one putative pathogenicity chromosome (red) and several other accessory sequences (dark blue). Accessory regions typically have (B) low gene density and (C) high repeat density, both calculated here in 50 kb windows. The putative pathogenicity chromosome is marked by (D) the presence of many candidate effector genes. (E) Indicates nucmer alignments with the Fol4287 reference assembly: in red alignments to the putative pathogenicity chromosome of Forc016, in dark blue alignments from known accessory regions in Fol4287 (chr1B; chr2B; chr3; chr6; chr14; chr15) and in light blue the remaining alignments, mostly between core regions in both genomes.
