Figure 3.
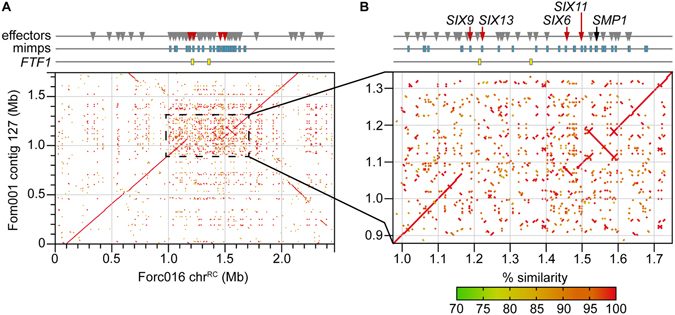
Comparison of chrRC with Fom001 contig 127 reveals a highly dynamic central region containing the majority of the miniature impala (mimp) TEs and candidate effector genes of the Forc genome. (A) The chromosomes of the Fom and Forc strains are mostly syntenic, with large stretches showing 100% identity in this nucmer alignment. (B) The dynamic central region of the chromosome, about 700 kb in size, has the highest repeat density, 30 of the 35 mimps of the Forc genome and the majority of the Forc candidate effector genes, including SIX9, 13, 6 and 11 and SMP1. Additionally, the two FTF1 homologs present in the Forc genome are found here.
