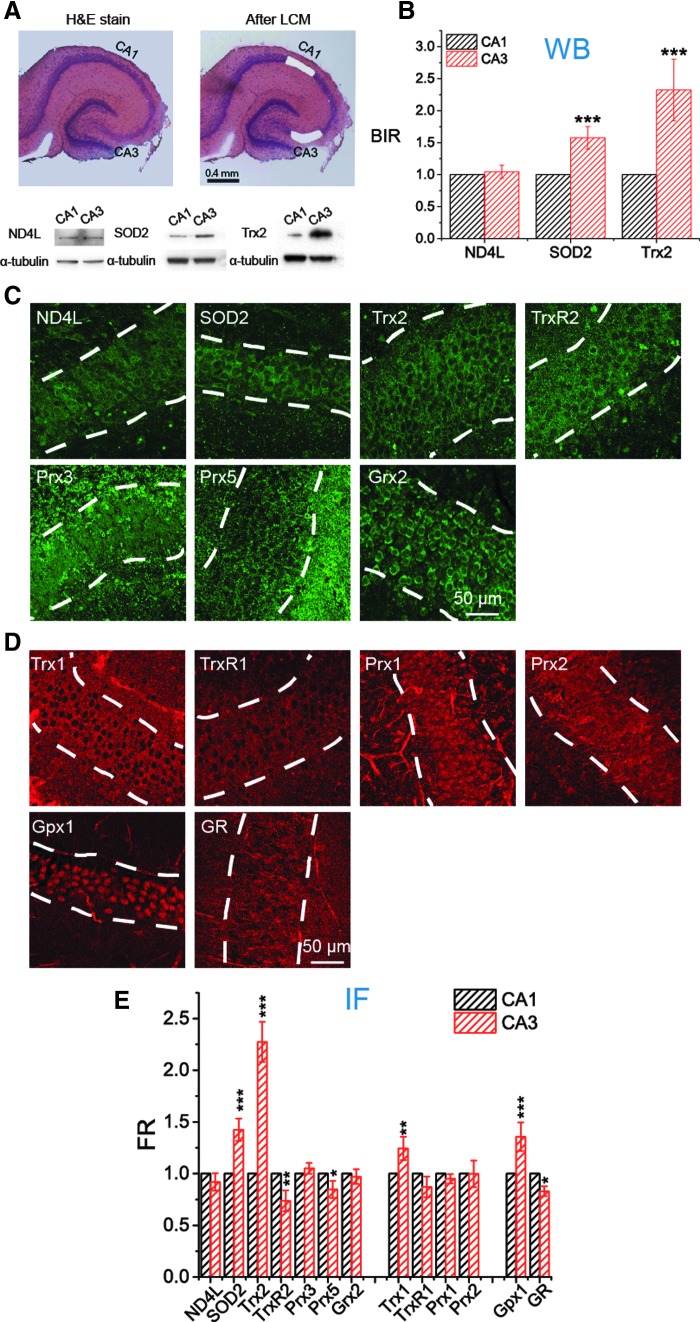
FIG. 4.
Expression of Trx2, Trx1, and SOD2 in pyramidal cells is significantly higher in CA3 than in CA1. (A, B) Comparison of the intensities of ND4L, SOD2, and Trx2 in CA1 and CA3 in WBs. (A) The pyramidal cell layers of CA1 and CA3 visualized by H&E staining were collected separately by LCM. For each individual WB, 50 LCM-generated bits of tissue were collected. Bottom, WB bands of ND4L, SOD2, and Trx2 from CA1 and CA3 subfields (see uncropped blots in Supplementary Fig. S10). (B) Expression of each protein from CA1 and CA3 is compared quantitatively (n = 3 repeats). BIR is used to refer the targeted protein to internal standard, α-tubulin. (C–E) Comparison of the contents of 13 proteins in CA1 and CA3 with IF. (C, D) Representative fluorescence images of subfield CA1 from immunostained OHSCs. The pyramidal cell layer is indicated by white dash lines. (C) Seven mitochondrial proteins are labeled with green fluorescent antibody shown in green. (D) First four proteins are mainly located in cytosol. GPx1 and GR exist both in mitochondria and cytosol. They are all labeled with red fluorescent antibody shown in red. (E) FR of the targeted protein (mitochondrial proteins referenced to MitoTracker, cytosolic proteins referenced to α-tubulin) is quantitatively analyzed and compared between CA1 and CA3 (n = 10 OHSCs, see Supplementary Figure S5 and Experimental Methods section in Supplementary Data for more details). Data of ND4L, SOD2, and Trx2 are consistent with that acquired with WB in (B). See Figure 1 legend for indicators of statistical significance. BIR, band intensity ratio; FR, fluorescence ratio; Gpx, glutathione peroxidase; GR, glutathione reductase; Grx, glutaredoxin; IF, immunofluorescence; LCM, laser capture microdissection; ND4L, NADH dehydrogenase subunit 4L; Prx, peroxiredoxin; SOD, superoxide dismutase; Trx, thioredoxin; TrxR, thioredoxin reductase; WB, Western blot.