Fig. 4.
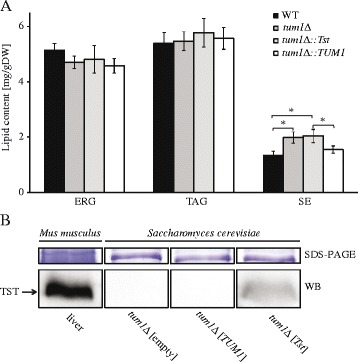
Yeast Tum1 and mouse Tst affect lipid metabolism in the Saccharomyces cerevisiae cells. a Lipid analysis of neutral lipids was performed by TLC. Three different groups of lipids were measured: ergosterol (ERG), triacylglycerols (TAG) and sterol esters (SE). The error bars represent standard deviation. Deletion of TUM1 causes a 60% increase of SE, as compared to the wild-type. This phenotype is also observed in the strain expressing Tst, indicating that Tst is not able to complement the loss of TUM1. b Western blot analysis confirms the presence of Mus musculus TST in the cells of Saccharomyces cerevisiae. Extracts of proteins were prepared and analysed by polyacrylamide gel electrophoresis and by western blotting with anti-TST antibodies. The Mus musculus liver protein sample was used as a positive control and tum1Δ and tum1Δ::TUM1 as negative controls. Coomassie Brilliant Blue staining of proteins in the gel with the same amount of protein as for the western blot analysis was used as a loading control. Boxes denote different parts of either the same gel (SDS-PAGE) or membrane (WB)
