Figure 1.
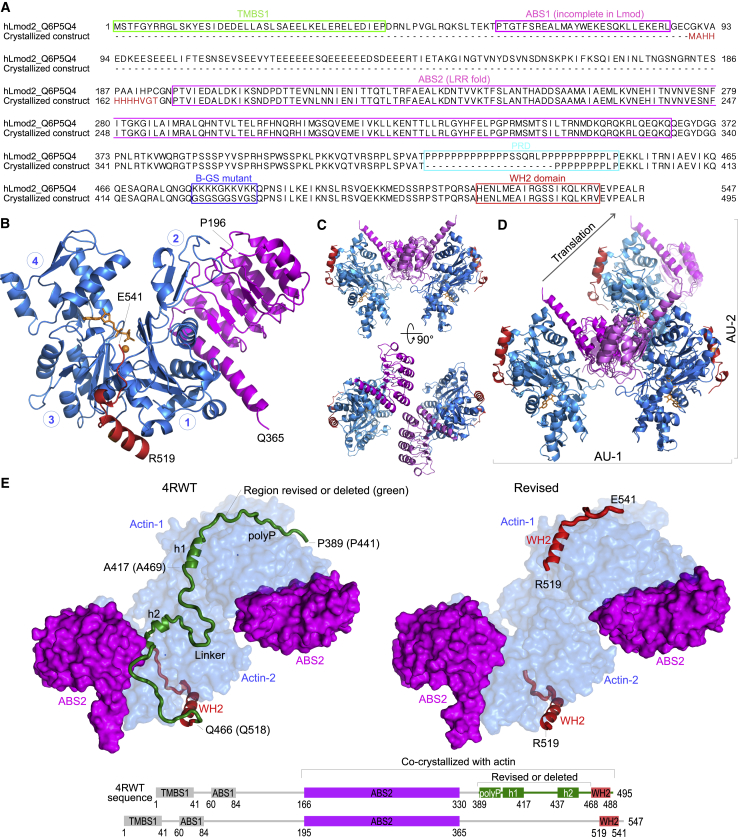
Revised structure of actin-Lmod2ABS2-C. (A) Shown here is an alignment of the correct sequence of human Lmod2 (UniProt ID: Q6P5Q4) and the Lmod2 construct cocrystallized with actin (8). The purification tag present in the crystallized Lmod2 construct is also shown at the N-terminus. Boxed regions correspond to the major Lmod2 subdomains implicated in interactions with actin (ABS1, ABS2, WH2 domain) and tropomyosin (TMBS1). The boxed region labeled “B-GS mutant” corresponds to a mutation introduced by Chen et al. (8) into the crystallized fragment to replace a basic patch prone to degradation by a Gly-Ser linker. (B) Given here is a ribbon representation of one of the two nearly identical actin-Lmod2 complexes present in the P1 unit cell. Circled numbers indicate the four subdomains of actin. The ABS2 and WH2 domain of Lmod2 correspond to the boxed regions shown in (A). ABS2 binds subdomains 1 and 2 of actin, whereas the WH2 domain binds at the barbed end of the same actin monomer, occupying the cleft between subdomains 1 and 3, known as the target-binding or hydrophobic cleft (20). The ATP analog AMP-PNP and associated Mg2+ ion in the nucleotide-binding cleft of actin are also shown. Terminal residues in ABS2 and the WH2 domain are labeled. (C) The P1 unit cell contains two copies of the actin-Lmod2ABS2-C complex, related by a local twofold symmetry axis, distinct from the helical symmetry of the actin filament. Two orthogonal views are shown. (D) Given here are two alternate but equivalent definitions of the contents of the asymmetric unit, AU, corresponding to the unit cell in the P1 space group. The AU-2 definition corresponds to that used by Chen et al. (8). AU-2 is the same as the twofold related complex (AU-1), but translated to an adjacent unit cell, as the arrow indicates. (E) Shown here is a comparison of the original (left) and revised (right) structures of actin-Lmod2ABS2-C according to the AU-2 definition. The region of the Lmod2 C-terminal extension present only in the original structure (residues P441–Q518, using the correct sequence numbering) is shown in the model and accompanying diagram and labeled as “revised or deleted”. The domains that are not present in the crystallized Lmod2 fragment or not visualized in the structures are shown in light gray in the diagram at the bottom, where both the original (top) and the correct (bottom) numbering schemes are shown for reference. The revised structure has two extra amino acids at the N-terminus of ABS2 and 83 fewer amino acids in the region between ABS2 and the WH2 domain, and also lacks the last six amino acids after the WH2 domain. Note also that there are two WH2 domains in the revised structure, whereas in the structure of Chen et al. (8), the position of the WH2 domains bound to one of actin molecules (labeled Actin-1) is occupied by a portion of the PRD (polyP) and helix h1, with reverse polypeptide directionality. To see this figure in color, go online.