Abstract
The insect pests are big threat in meeting the food demands for future generation. The present pest control strategies, including the existing transgenic approaches show certain limitations and are not completely successful in limiting the insect pests. However, the sequence-specific gene silencing via RNA interference (RNAi) holds a great promise for effective management of agricultural pests. RNAi is naturally occurring conserved process responsible for gene regulation and defense against pathogens. The efficacy of RNAi varies among different insect orders and also depends upon various factors, including the target gene selection, method of dsRNAs delivery, expression of dsRNAs and presence of off-target effects. RNAi-mediated silencing of different insect genes involved in various physiological processes was found to be detrimental to insects growth, development and survival. In this article, we have reviewed the potential of RNAi-based strategies for effective management of insect pests. We have also discussed the various parameters, which are to be considered for host-induced RNAi-mediated control of insect pests without producing any effect on non-target organisms and environment.
Keywords: Insect pest management, Transgenic plants, RNA interference, Gene silencing, Biotic stress
Introduction
The insect pests are important constraint in achieving the global food demands. The potential yields of all agricultural crops are affected substantially due to direct or indirect effects of insect pests. Direct damage includes fouling, deformations or necrosis of plant tissues or organs and dissemination of plant pathogens, whereas indirect damage involves the loss of harvest quality (in term of damaged fruits) and increase in overall cost of crop production (Bardner and Fletcher 1974). The estimated overall annual yield loss due to insect pests in major crops account approximately 18% in the absence of control measures (Oerke 2006). Worldwide, insects are the major pests of a wide range of crops including cotton, chickpea, pigeon pea, tomato, maize, groundnut, sunflower and tobacco (Lal 1985; Mehto et al. 1985). Only 0.5% of insects produce serious menace to humans and are given the status of ‘Pest’. Ecological and physiological characteristics of insects, include a tough exoskeleton, small body size, ability to fly, a high reproductive potential and adaptability in an ever-changing environment, polyphagy nature, ability to facultative diapause, inherent capability of evolving to resistant biotypes contributes to successful establishment of the insects as pest (Roush and McKenzie 1987). Most of the agricultural pests belong to Lepidopteran, and there is hardly any cultivated plant, which is spared by these pests. Some economically important genera of lepidopteran insects are listed in Table 1.
Table 1.
List of major lepidopteron agricultural pests.
Source Puri and Ramamurthy (2009)
| Insect pest | Scientific name | Crop(s) |
|---|---|---|
| American bollworm | Helicoverpa armigera (Hubner) | Cotton, chickpea, pigeonpea, sunflower, tomato |
| Whitefly | Bemisia tabaci (Gennadius) | Cotton, tobacco |
| Brown planthopper | Nilaparvata lugens (Stal) | Rice |
| Green leafhopper | Nephotettix spp. | Rice |
| Serpentine leaf miner | Liriomyza trifolii (Burgess) | Cotton, tomato, cucurbits, several other vegetables |
| Fruit fly | Bactrocera spp. | Fruits and vegetables |
| Mealy bugs | Several species | Several field and horticultural crops |
| Thrips | Several species | Groundnut, cotton, chillies, roses, grapes, citrus and pomegranate |
| Wheat aphid | Macrosiphum miscanthi (Takahashi) | Wheat, barley, oats |
| Pink stem borer | Sesamia inferens (Walker) | Wheat |
| Gall midge | Orseolia oryzae (Wood-Mason) | Rice |
| Gall midge | Several species | Fruit crops |
| Diamondback moth | Plutella xylostella (Linnaeus) | Cabbage |
| Hoppers | Several species | Mango |
| Pyrilla | Pyrilla perpusilla (Walker) | Sugarcane or rice at times |
| Polyphagous pests like termites, white grubs, hairy caterpillars and tobacco caterpillar | Several species | Many agroecosystems |
Current pest control strategies and their limitations
Currently, different strategies such as cultural, mechanical, biological, chemical and transgenic approaches are utilized for effective control of insect pests. Of these, the cultural, mechanical and biological measures are traditional approaches, being followed by farmers since ages. The cultural methods of control involve proper cropping methods, crop rotation, land, water and post-harvest management. Mechanical methods include picking and trapping of insects. Use of parasites, parasitoids and predators comprise the biological methods. These methods are environment-friendly, self-perpetuating, cost-effective, easily available, convenient to use, pose no harmful effects on environment, and are being compatible with other strategies. However, they are generally slow in action, require skilled personnel, and can be applied to only a small area at a given time. Chemical methods involve use of toxic substances, which interfere with one or more vital pathways through inhibition of enzyme activities (Stenersen 2004). Chemical methods are the quicker and more effective as compared to other methods of pest control. Every year, a large sum of money is invested on crop protection through the use of chemical insecticides. The main disadvantages associated with chemical methods are their persistence and biomagnifications in the environment, which causes environmental as well as health related problems. Plant breeding approaches include the introgression of insect resistance trait into the variety of interest (Yencho et al. 2000). However, plant breeding is limited by lack of resistance in the germplasm for many pests and introgression of undesirable harmful traits from the wild varieties through linkage drag.
Therefore, it is imperative to look for novel and effective alternative approaches to develop crop resistance against insect pests. In recent years, biotechnology has provided additional tools to limit the damages caused by insect pests while at the same time has given solutions against the limitations of traditional and hazardous chemical methods (Christou et al. 2006; Huang et al. 2002). Transgenic approaches offer many advantages over the above mentioned traditional methods of pest control (Hilder and Boulter 1999; Sharma et al. 2002; Sharma and Sharma 2013). They are more specific in their action against insect pest and produce insecticidal compounds continuously in large amount in transgenic plants. Thus, they are considered more economical to farmers in term of increased crop yield. In 1987, first tobacco transgenic plant expressing cowpea trypsin inhibitor protein was developed against Heliothis virescen. Cowpea trypsin inhibitor was found less effective against various insect pests. Therefore, search for more potent insecticidal proteins was initiated. Insecticidal proteins such as Bacillus thuringiensis endotoxin, plant protease inhibitors (PIs) and alpha-amylase inhibitors, chitinases, lectins and biotin binding proteins, secondary metabolites, isopently transferases and vegetative insecticidal proteins (VIPs) from various sources, including bacteria, plants and insects were exploited for generation of insect resistant transgenic plants (Stevens et al. 2012). Bt endotoxin expressing transgenic crop plants has been commercially very successful in controlling the insect pests so far (Christou et al. 2006) and has significantly reduced the use of chemical pesticides (James 2014). However, the rapid evolution of resistance by insect pests against insecticidal toxin (Tabashnik et al. 2013), ineffectiveness of toxin against various pests, effect on non-targeted organisms and on the micro-environment are the major drawbacks of existing transgenic approaches. The insect species gain resistance against insecticidal proteins due to sub-optimal expression of toxin, mutation in the target gene of insect pest, loss of target midgut protease due to selection pressure, over-expression of sensitive protease, synthesis of insensitive proteases and change in the membrane integrity. Considering of the current situation of crop protection, it is necessary to analyze the characteristics of an ideal pest control strategy (Hilder and Boulter 1999). The ideal pest control strategy should be economical, environmental and farmer friendly. It should be specific in its action and should target large number of pests without affecting non-targeted organisms. The technology should have an alternative way, in case of development of resistance by the pest. In order to achieve the ideal pest control method, there is an urgent need for exploring other possible approaches for imparting broad spectrum insect resistance.
RNAi: next generation pest control strategy
RNAi silencing in insects
According to the central dogma, RNA is just a bypass of genetic information from DNA to protein but after the discovery of ‘antisense-mediated silencing’ of homologous genes, its role in regulating gene expression was well established (Nellen and Lichtenstein 1993). Fire et al. (1998) proved that double-stranded RNA (dsRNA) is the predominant trigger of gene silencing, as compared to sense or antisense RNA alone in Caenorhabditis elegans. This breakthrough discovery led to the establishment of the rapidly growing field in the biological world named as RNA interference (RNAi). Thus, RNAi is defined as sequence-specific silencing of target gene. A similar homology-dependent gene silencing phenomena was found in plants known as ‘co-suppression or post transcriptional gene silencing’ and in fungi as ‘quelling’ (Cogoni et al. 1996; Napoli et al. 1990; Romano and Macino 1992). Bacteria also showed RNAi like phenomenon, known as clustered regularly interspaced short palindromic repeats (CRISPR), (Wilson and Doudna 2013). RNAi and related pathways are evolutionarily conserved cellular process. They are required in regulation of gene expression, genomic re-arrangements and defense against foreign nucleic acid (Carthew and Sontheimer 2009; Perrimon et al. 2010). RNAi pathway involves formation of interfering molecules through activity of dicer enzyme. These interfering molecules can be small interfering RNAs (siRNAs) and microRNAs (miRNAs). The interfering molecules are then loaded onto RNA induced silencing complex (RISC) comprising of Argonaute protein (AGO). RISC directs the interfering molecules to their cognate target where homology based cleavage of target mRNA occurs (Wilson and Doudna 2013). RNAi is more efficient, user friendly, flexible, specific and stable technique for pest control. It has been exploited successfully as a powerful reverse genetic tool to study the function of genes and biological control of various agricultural insect pests and pathogens (Andrade and Hunter 2016; Belles 2010; Gordon and Waterhouse 2007; Palli 2014; Price and Gatehouse 2008; Saurabh et al. 2014; Thakur et al. 2016; Yogindran and Rajam 2015; Zotti and Smagghe, 2015).
Systemic RNAi in insects
RNA-dependent RNA polymerase (RdRP) mediated amplification of siRNA molecules were reported in plants, nematodes and fungi, but was found to be absent in humans and insects (Gordon and Waterhouse 2007). Surprisingly, Tribolium castaneum also exhibited robust systemic RNAi response even in the absence of RdRP (Tomoyasu et al. 2008). Insect genomes do not have homologues of canonical RdRPs (Gordon and Waterhouse 2007; Jose and Hunter 2007; Richards et al. 2008), but a subunit of the RNA polymerase II has been identified in Drosophila melanogaster, which possesses similar activity as that of RdRP and involved in RNAi and transposon suppression in D. melanogaster (Lipardi and Paterson 2009). The presence of trans-developmental and trans-generation effects of RNAi in some insects further proves the systemic nature of RNAi in insects. Based on these results, it was postulated that the amplification of RNAi in insects might be due to the presence of some unknown mechanisms analogous to RdRP.
Cellular up-take of dsRNA in insects
RNAi signals found to be mobile within organism and among organisms belong to different kingdoms. The movement of siRNA signal across kingdom has been observed in wide range of organisms from bacteria to nematodes, plants to nematodes, fungal pathogens and insect pests, and human to insect parasites (Knip et al. 2014). Insects can take up siRNAs directly from the environment or tissue and transfer signal from cell to cell. The ability of execution of RNAi by single cell is termed as cell autonomous RNAi. In this case, the site of production or introduction of dsRNAs and its RNAi effects is same, whereas in non-cell autonomous RNAi, the site of RNAi effect is different from the site of dsRNA production or introduction. Non-cell autonomous RNAi can be environmental, when cell takes the dsRNA molecules from environment or systemic RNAi, when cell takes the dsRNA molecules from other cell or tissue and spread these siRNA molecules to other parts (Baum and Roberts 2014; Huvenne and Smagghe 2010). A robust silencing response in insects was observed to both environmental and systemic RNAi (Prentice et al. 2015; Price and Gatehouse 2008; Tomoyasu et al. 2008). Two pathways have been proposed to explain the systemic silencing in insects, trans-membrane channel-mediated uptake mechanism and an alternative endocytosis-mediated uptake mechanism (Huvenne and Smagghe 2010; Joga et al. 2016; Xue et al. 2012).
The trans-membrane channel-mediated uptake mechanism involves two trans-membrane proteins, named as SID-1 (systemic RNAi defective) and SID-2 (Cappelle et al. 2016; Jose et al. 2009). SID-1 is a multispan trans-membrane protein, found in all non-neuronal cells (Winston et al. 2002). It transports dsRNA passively among the C. elegans cells (Cappelle et al. 2016; Jose et al. 2009). The other protein, SID-2 is directly involved in the uptake of ingested dsRNAs and is expressed in the worm intestine tissue (Cappelle et al. 2016). Thus, non-autonomous RNAi involves both SID-1 and SID-2 functions sequentially. SID-2 mediates the initial uptake of dsRNA directly from the intestinal lumen or environment, while SID-1 functions at secondary step and transports the dsRNAs into the cytoplasm (Cappelle et al. 2016; Jose et al. 2009). Similar process is also observed in other metazoans which is associated with selective import of extracellular dsRNA. In silico analysis of Sid-1 homologs of T. castaneum showed that Sid-1 homologs have more similarity with C. elegans tag-130, as compared to Sid-1. It was also found that, tag-130 gene was not involved in systemic RNAi in C. elegans (Tomoyasu et al. 2008). Thus, SID-1 is not imperative for uptake of silencing signal in insects. However, an alternative dsRNA uptake mechanism might exist in insects, since robust systemic RNAi response was observed in some insects such as T. castaneum and mosquitoes even in the absence of Sid orthologs (Boisson et al. 2006; Tomoyasu et al. 2008).
The endocytosis-mediated silencing signal uptake mechanism was based on the receptor-mediated endocytosis (Saleh et al. 2006; Ulvila et al. 2006). According to this model, insect cell takes the silencing signal from environment by receptor-mediated endocytosis and then actively spreads the silencing signal through vesicle-mediated intracellular trafficking (Tomoyasu et al. 2008; Saleh et al. 2006). The receptor mediated uptake mechanism involves secretion of siRNA signal after its production in the form of vesicle, uptake of siRNAs through receptor for execution of siRNA-mediated silencing (Knip et al. 2014). When S2 cell of D. melanogaster was used to study the uptake mechanism, it was found that more than 90% of dsRNA uptake depends on SR-CI and Eater receptors. The D. melanogaster SR–CI shows similarity with the mammalian class A scavenger receptors, suggesting the possibility of involvement of receptor-mediated endocytosis in the uptake of dsRNA in D. melanogaster (Saleh et al. 2006; Ulvila et al. 2006). Abolishment of RNAi phenomenon in C. elegans was observed due to mutation in endocytosis-mediated genes, indicating the evolutionary conservation of endocytosis-mediated dsRNA uptake mechanism in insects (Saleh et al. 2006). However, the mechanism by which dsRNA are imported to the suitable position in the cell through endocytosis was not clearly understood. It was proposed that different receptors, adapters and sorting signals might play an important role in this process (Saleh et al. 2006). Recently, uptake through Sid-1-like channel proteins and receptor-mediated endocytosis was observed in Colorado potato beetle, Leptinotarsa decemlineata (Cappelle et al. 2016).
Challenges for successful RNAi in insects
Despite the tremendous utility of RNAi as a promising strategy for studying fundamental biological questions and for control of insect pests, there is still a need to analyze several aspects of RNAi before establishing it as a long-term effective pest control method in the field. RNAi application and efficacy remains variable among different genes, tissues, organisms and life stage of insect. For example, RNAi effect has been found to be more in hemocytes of D. melanogaster and Manduca sexta as compared to other tissues when injected with target gene dsRNAs (Mao and Zeng 2014; Miller et al. 2008). The success of RNAi experiments in different species was also influenced by many biological variables such as variation in the core RNAi machinery, cellular uptake and propagation of silencing signal and dsRNA degrading enzymes, as well as other differences in genetic backgrounds (Kitzmann et al. 2013; Miller et al. 2008). The aforementioned challenges can be pacified by considering different experimental factors during designing of experiment, which include the mode of delivery, dose of the dsRNA molecule and target gene. We are describing few potential challenges involved in using RNAi as a crop protection strategy in the following sections.
Digestion of dsRNA by insect gut nucleases
Nucleic acid degrading enzymes found inside the insects gut form an integral part of the digestive cocktail of insects. The dsRNA molecules are potent substrate for these nucleases inside the gut and can be easily degraded by them. Therefore, protection of the ingested dsRNA from the action of nuclease is necessary for initiation and functionality of whole RNAi mechanism. Very little is known about fate of dsRNA inside the insect gut after the ingestion of plant material. In tarnished plant bug, Lygus lineolaris, delivery of polygalacturonase dsRNA by injection led to down-regulation of gene, while dsRNA intake by feeding approach did not produce any response. The decrease in the RNAi response was also observed due to the presence of nucleases in the insect saliva and midgut (Allen and Walker 2012; Garbutt et al. 2013; Wynant et al. 2014). Garbutt et al. (2013) explained the differential response of two insects to RNAi by studying the persistence of dsRNA in their hemolymph. They found that nucleases rapidly degrade dsRNA in M. sexta hemolymph plasma, while dsRNA persisted for much longer in B. germanica plasma. The studies proposed that the susceptibility of insect species to RNAi can be defined by the rate of persistence of dsRNA molecules in the hemolymph (Garbutt et al. 2013).
Chemical hydrolysis of dsRNA by insects gut pH
Gut pH through the activation and deactivation of certain set of enzymes digests food material in the insect gut. Due to the change in pH along the gut from acidic in the anterior midgut (AM) to basic in posterior midgut (PM), restricted enzyme activity in different areas of the larval midgut was seen (Vinokurov et al. 2006). Chemical hydrolysis (increases with increasing pH) and enzymes in the gut alone or both together, could affect the stability of dsRNA (Hakim et al. 2010). Thus, protection of dsRNA is required from the hostile environment of insect gut. Coating of dsRNA molecules was one of such approach which protects the dsRNA from nucleases and pH variation (Huvenne and Smagghe 2010). Lepidopteran insects showed less efficient RNAi as compared to coleopteran insects due to difference in hemolymph composition, uptake and processing of dsRNA mechanism. It was observed that dsRNA degraded with fast rate in hemolymph of lepidopteran insect in comparison to coleopteran hemolymph (Shukla et al. 2016). Thus, difference in gut environment contributes to variation in RNAi efficiency in insects. Some insects showed robust RNAi response even in the hostile environment of insect gut, which might be due to the presence of associated factors required for the stability of dsRNAs inside the insects gut (Belles 2010; Kola et al. 2015; Xu et al. 2016).
Amount of dsRNA molecules
Insect species, life stage, delivery method, abundance of the target gene transcript and its spatial and temporal expression profiles decide the requisite amount of dsRNA molecules for optimal silencing. The insect internal factors such as the mode of uptake and ability to spread RNAi molecules also strongly influence the optimal dose of dsRNA, e.g. in D. melanogaster, RNAi cannot be induced through extracellular injection in tissues other than hemocytes, due to the absence of dsRNA uptake machinery (Miller et al. 2008). Multiple introductions of dsRNA molecules can reduce or enhance the RNAi efficiency (Araujo et al. 2006; Shakesby et al. 2009), for example salivary glands of Rhodniu prolixus showed enhanced RNAi response when supplied with high or mixed dose of dsRNA molecules (Araujo et al. 2006). The basis of enhanced RNAi response in this organism can be due to the activation of RNAi machinery by high dose of dsRNA molecules (Garbutt et al. 2013; Liu et al. 2013). Whereas, the high or mixed dose of dsRNA leads to competition between dsRNA molecules for RNAi machinery and results in oversaturation of RNAi machinery components, thereby reducing the RNAi efficiency (Miller et al. 2012). By optimizing the concentration of dsRNA for orthologouse genes, off-target effects can be minimized, e.g. silencing of vacuolar H+ ATPase gene in Leptinotarsa. decemlineata and D. virgifera virgifera by a single dsRNA depends upon the dose of dsRNA molecules (Baum et al. 2007).
Length of dsRNA molecules
The length of dsRNA utilized to produce siRNA molecules determined the uptake and silencing efficiency of RNAi in an organism (Mao et al. 2007; Saleh et al. 2006). The minimal length of dsRNA, required to obtain maximum RNAi silencing, varies among insect species (Bolognesi et al. 2012). In most of the insect feeding experiments, sequences ranging from >50 to 500 bp were used to obtain greater success with RNAi (Andrade and Hunter 2016; Huvenne and Smagghe 2010). Long, unprocessed dsRNA fragments have been more effective in silencing of genes as compared to siRNA produced by in vitro synthesis or Dicer activity (Mao et al. 2007). Jin et al. (2015) expressed the dsRNA in chloroplast and showed efficient down-regulation of target genes. Chloroplast lacks dsRNA processing machinery, the unprocessed dsRNAs were taken directly by insect for induction of RNAi pathways (Jin et al. 2015). Recent report showed that processing of dsRNA in fungi after uptake produced prominent silencing effect as compared to processed siRNA uptake in Fusarium graminearum (Koch et al. 2016). The longer dsRNAs greater than 200 bp will produce more siRNA molecules through dicer activity, which can be easily taken up by uptake machinery (Andrade and Hunter 2016; Miller et al. 2012). Contrastingly, there are a few reports where a single chemically synthesized siRNA molecule were successfully used to silence acetylcholinesterase (AChE) gene in H. armigera and tsetse fly (Attardo et al. 2012; Kumar et al. 2009). Thus, long and short dsRNAs are effective in inducing gene silencing depending upon the target pest and target gene.
Life stage of insects
Most insects show prominent RNAi when targeted at younger stages, due to their smaller size and less developed body, for example Rhodnius prolixus showed efficient RNAi response, when nitropin 2 was silenced at second instar stage as compared to its silencing at 4th instar stage with same dose of dsRNA (Araujo et al. 2006). Similarly S. frugiperda, 5th instar larvae showed more silencing effect as compared to the adults moths (Vinokurov et al. 2006). Parental RNAi was more prominent when the female pupae or adults were targeted with dsRNA as compared to the last instar stage (Bucher et al. 2002). The difference in the RNAi efficiency at different insect stages was contributed by the differences in physiological as well as genetic characteristics of insects.
Mode of dsRNA delivery methods
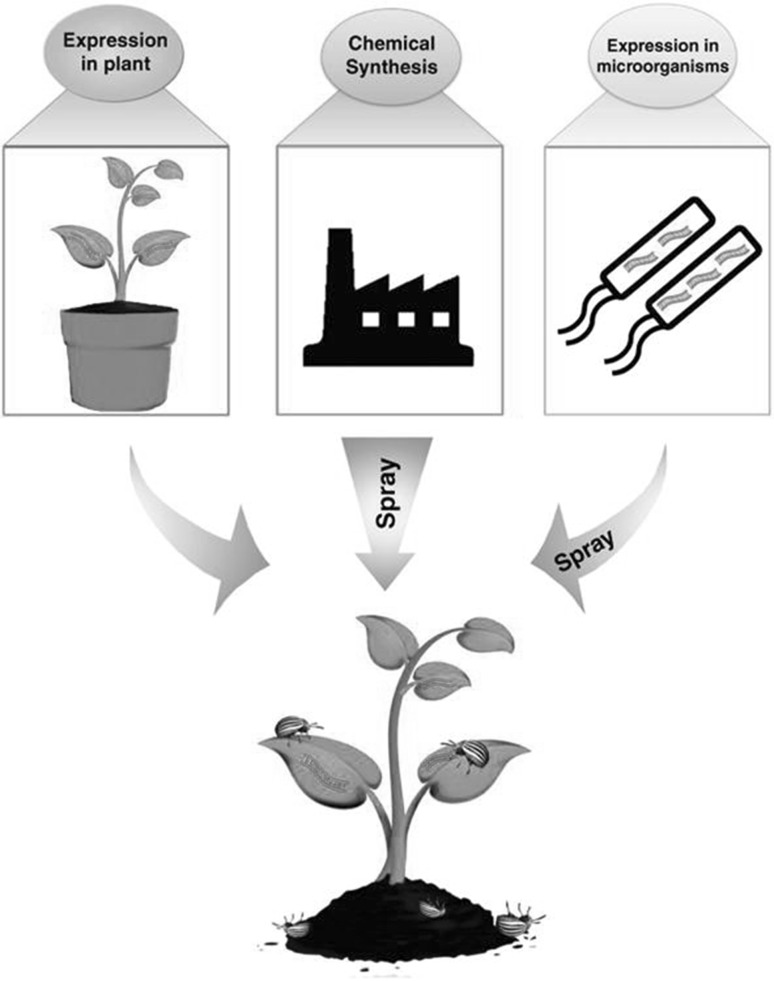
A successful RNAi response can be achieved by using efficient delivery method. The major dsRNA delivery methods explored till now in different organisms include soaking, microinjection and feeding. Production of interfering molecules at mass scale through these methods can be used as insecticides (Fig. 1).
Fig. 1.
Use of RNAi as a future insecticide
(Source adopted from Palli 2014)
Soaking or incubation
Soaking is one the best methods to study RNAi effect in cell culture. In this delivery method, cells or tissues are soaked in dsRNA solution for a particular time. Transfecting agents increase the RNAi efficacy by assisting the uptake of dsRNA from the solution. Uptake of dsRNA through soaking was first reported in C. elegans by Tabara et al. (1998). Since then, this delivery method was explored in different organisms such as D. melanogaster, nematodes and flatworms (Orii et al. 2003; Tabara et al. 1998). But, due to its limited applicability, it is rarely used in different experiments.
Injection
Simple protocol and its effectiveness in silencing the gene expression made microinjection the most popular dsRNA delivery method. In this method, dsRNA is injected into the target tissue or hemolymph directly with the help of proper inserting needle. Fire et al. (1998) were the first to use microinjection in their experiments on C. elegans for evaluation of the effects of sense and antisense RNA, singly and in conjunction. Till now, a wide range of insect species have been tested through dsRNA injection for functional genomics studies, also proving the utility of RNAi for elucidating the gene functions in evolutionarily divergent organisms (Belles 2010; Liu et al. 2010; Ulrich et al. 2015). The major advantage of using the microinjection as the delivery method is that the dsRNAs can be directly introduced into the targeted tissue with the known concentration though it is an expensive technique, requires prior optimization and highly skilled personnel, not feasible in field (Gu and Knipple 2013; Xue et al. 2012; Yu et al. 2013). Therefore, this delivery method has very limited application as pest control strategy.
Spraying
In this method dsRNA or siRNA are synthesized in vitro using different methods and then spray onto the plant surface. Earlier it was thought that spraying will leads to degradation of siRNAs but recent reports suggest that it can be utilized as an effective delivery method for dsRNA/siRNA. Miguel and Scott (2016) sprayed dsRNA of actin gene on leaves against Colorado potato beetle. They found that dsRNA sprayed plants remained protective against pest for 28 days under green-house conditions and dsRNAs were stable on leaf once they got dried. A similar method of delivery was also found effective against fungal pathogens (Koch et al. 2016) and viral pathogen (Konakalla et al. 2016). Spraying requires production of dsRNA or siRNA at large scale, which makes this approach costly otherwise it is an easy way of dsRNA or siRNA delivery to plant without undergoing any time taking approaches. Phloem sap feeding and stem borer insect pest cannot be targeted through spraying as these pest feed on internal sap as compared to surface (Li et al. 2015).
Feeding
The delivery of dsRNA by oral feeding is less invasive and comparatively simple as compared to injection. Timmons and Fire (1998) demonstrated for the first time the delivery of dsRNA molecules through oral route. Till now, this approach has been successfully practiced in seven different insect orders, including at least 15 different species ranged from agricultural pests to human parasites (Huvenne and Smagghe 2010). Different dsRNA delivery methods through feeding approach include the feeding of dsRNA expressed in bacteria and chemically synthesized dsRNA through the artificial diet, Nanoparticle/liposome-mediated dsRNA feeding and feeding of dsRNA through expression in transgenic plants (Joga et al. 2016; Mamta et al. 2016; Xue et al. 2012). So far, in most of the functional genomics studies, an artificial diet based feeding approach was used to decipher the function of genes (Araujo et al. 2006; Tian et al. 2009; Walshe et al. 2009). In this method, the artificial diet is mixed with dsRNA/engineered bacteria and the insect was made to feed on it. For example, dsRNA feeding of chitinase gene on artificial diet caused mortality and growth retardation in Mythimna separate (Ganbaatar et al. 2017). In addition, bacterial feeding also provides an alternative method for large-scale screening for target candidate genes (Mao et al. 2007).
In liposome or nanoparticle based delivery system-dsRNAs containing nanoparticles/liposome are generated and then, these particles are delivered to insect through feeding on artificial diet (Zhang et al. 2010). Nanoparticles/liposome stabilizes the dsRNA molecules during delivery process and thereby, increases the efficiency of RNAi. Liposomes are generally formed by conjugation of lipophilic molecules such as cholesterol, bile acids, and long-chain fatty acids (Batzri and Korn 1973). Liposomes-mediated efficient uptake of siRNA molecules and silencing response was observed in mice and D. melanogaster (Whyard et al. 2009; Wolfrum et al. 2007). Similarly, in nanoparticle based delivery method, dsRNAs are made to entrap into chitosan polymer via electrostatic forces to form a chitosan/dsRNA nanoparticles (Howard et al. 2006; Zhang et al. 2010). Recently, approximately 40–50% liposome-mediated silencing was observed in spotted wing Drosophila (Taning et al. 2016). However, chitosan/dsRNA based nanoparticles delivery methods for RNAi has not been fully utilized for insect pest control. To make RNAi effective against large number of herbivore insect pests, plants should be engineering to produced dsRNA against the insect vital genes. Generally, stable transformation by RNAi vector is employed for generation of transgenic plants expressing dsRNA. Transgenic plants are generated by using RNAi vector which contains the target gene in sense and antisense orientation separated by an intron (spacer). The dsRNAs/siRNAs formed inside the transgenic plants are taken up by insects upon feeding, which lead to the silencing of the target gene in insects. Generation of transgenic plants expressing dsRNA is a more practical method as a pest control strategy for field applications as compared to other delivery methods. It is labor-intensive, cost-effective and easy to perform. The disadvantage associated with this method is that a prior optimization is required as the exact amount of dsRNA ingested is difficult to measure (Burand and Hunter 2013; Xue et al. 2012; Zhang et al. 2013). Plants have RdRp for amplification of RNAi signals and SID for transport of RNAi signal, whereas RdRp and SID are absent in insects. Thus, plants amplify the siRNA molecules and transport them through phloem and plasmodesmata to other parts. Insect takes processed and unprocessed dsRNAs from plants through feeding and then move dsRNA to other body parts or target sites. Robust and systemic RNAi response was observed in insects even in the absence of RdRp and SID-1, which might be due to the presence of other genes or pathways required for the amplification and systemic response.
Target gene selection
The outcome of RNAi effects in the insect mainly depends upon the selection of an ideal target gene. The ideal gene target for RNAi should be vital for insect survival, and must be highly expresses. It should not have functional redundancy, so that the silencing effect can be seen easily (Li et al. 2013; Lomazzo et al. 2011). Terenius et al. (2011) summarized the response of lepidopteran insects to RNAi and found that out of 130 genes, 50 genes showed robust RNAi. Thus, variation in RNAi response of different genes will depend upon the vitality of the target gene to insect survival and redundancy of gene in its function. Longer self-life of protein/mRNA also contributes to the weak RNAi responses, e.g. silencing of nicotinic acetylcholine receptor subunit (Da6) gave a very low RNAi response in both D. melanogaster and T. castaneum due to the long shelf-life of its protein (Rinkevich and Scott 2013).
Off-target effect of the target gene
Off-target effect is one of the major limitation associated RNAi technology. Off-target effects are describes as the silencing of non-target genes in the same organism or in non-target organisms. Sequence homology of siRNAs to the non-target genes especially in its 3′ UTR region can cause off-target effects (Birmingham et al. 2006; Jonathan and Jian 2013). However, this limitation can be partially overcome by specific selection of the target region.
The specificity of RNAi effect depends upon the selection of gene region, which is intended to produce dsRNA molecules. Through designing dsRNA sequence from a very less conserved gene region, species-specific silencing of highly conserved gene can be achieved. Whyard et al. (2009) showed that the V-ATPase genes of T. castaneum, M. sexta, A. pisum and D. melanogaster can be specifically silenced without adversely affecting the other species using species-specific dsRNA. Thus, off-target effects can be minimized or prevented by using species-specific dsRNA, or expressing dsRNA in inducible and tissue-specific manner Different web-based computational tools are freely available for designing of the off-target free RNAi constructs. The above mention results prove that, RNAi can be used for effective silencing of specific targets as well as for testing the potential impact of insecticidal RNAs to non-target organisms.
Host-induced RNAi for insect pest control
RNAi can be successfully employed as a control strategy against insect pests (Gordon and Waterhouse 2007; Huvenne and Smagghe 2010; Joga et al. 2016). In most of functional genomic studies, injection or feeding of bacteria expressing the dsRNA was used as a delivery method to silence different target genes. Silencing of some genes had produced devastating effects on the insect growth, development, and survival (Xu et al. 2016). These studies suggested the possibility of utilization of feeding bioassay approach for control of insect pests through RNAi.
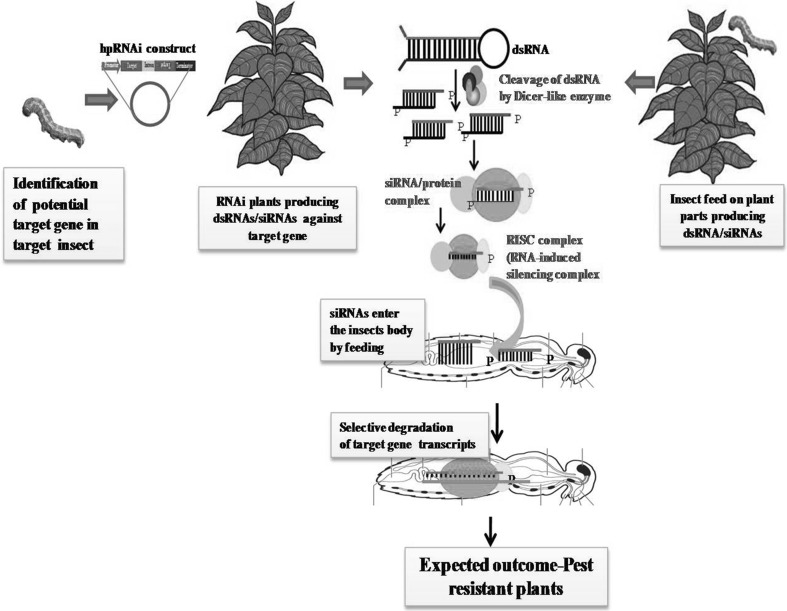
In plant-mediated or host-induced RNAi (HI-RNAi) approach, a crop plant is engineered with hair-pin RNAi vector to produce dsRNA against the target gene of insect pest. Upon feeding on plant parts, dsRNA enters into the insect gut, leading to the induction of RNAi machinery and then, silencing of the target gene in the insect pest (Fig. 2). The success of HI-RNAi was first demonstrated by Baum et al. (2007) and Mao et al. (2007) in their studies on Diabrotica virgifera virgifera LeConte and Helicoverpa armigera respectively. The dsRNAs were expressed in Nicotiana tobacum against H. armigera cytochrome P450 (CYP6AE14) gene (Mao et al. 2007). CYP6AE14 gene is involved in detoxification of gossypol and highly expressed in the midgut. Feeding of H. armigera larvae on dsRNA expressing CYP6AE14 N. tobacum plants caused reduction in CYP6AE14 transcripts. As a result, larvae were not able to detoxify gossypol and showed retarded growth (Mao et al. 2007). Baum et al. (2007) through feeding of dsRNA on artificial diet tested over 290 genes from Western corn rootworm (WCR) cDNA library in order to identify potential target genes. They found 14 potential genes and among them, vacuolar ATPase subunit A (V-ATPase) gene was selected for detailed analysis through HI-RNAi. They generated corn transgenic plants expressing dsRNA against the V-ATPase gene of WCR and observed that transgenic corn plants showed significantly less root damage as compared to the control plants in feeding bioassays (Baum et al. 2007). Later, HI-RNAi was also used for control of different insects through silencing of various insect vital genes (Jin et al. 2015; Mamta et al. 2016; Thakur et al. 2014; Xiong et al. 2013; Yogindran and Rajam 2015; Zhu et al. 2012). Hi-RNAi was also combined with other transgenic approaches in order to enhance resistance against pests. For instance, transgenic cotton plants expressing both dsCYP6AE14 and 35GhCP1 (Gossypium hirsutum cysteine protease) were found to be highly resistant to cotton bollworm than either of the single- transgenic lines (Mao et al. 2013). HI-RNAi also gave great advantage in controlling the sap sucking insects. Sap sucking insects were found insensitive towards Bt toxin. They pose great threat by acting as a vector for many virus born diseases (Li et al. 2011; Malik et al. 2016; Price and Gatehouse 2008). Mao and Zeng (2014) showed that the HI-RNAi silencing of gap gene in Myzus persicae impaired the reproductive potential of insect through reduction in target gene transcripts. The various studies on HI-RNAi for control of insect pests are summarized in Table 2.
Fig. 2.
Host-induced RNAi strategy for insect pest control
Table 2.
Summary of HI-RNAi for the control of insect pests.
Source adopted and modified from Navale et al. (2014)
| Insect order | Target insect | Target genes | Functions | Gene fragment | Location | Intron | Binary vector | RNAi plant M | Effect of HI-RNAi on the target insect | Off-target effects | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lepidoptera | Helicoverpa armigera | Cytochrome 450 monooxygenase CTP6AE14 | Degradegossypol and increase tolerance to gossypol | 448 bp | Midgut | Arabidopsis RTM gene | pBI121 | Arabidopsis thaliana | Enhance gossypol toxicity in larvae, decreased larval weight and size with decrease in target gene transcripts | No off-target effects | Mao et al. (2007) |
| GST glutathion transferase | Detoxif ication enzymes | 448 bp | Whole insect body | Arabidopsis RTM gene | pBI121 | Tobacco | Depletion in transcript level, reduction in larval weight and size | No off-target effects | Mao et al. (2007) | ||
| Cytochrome 450 monooxygenase CTP6AE14 | Degradegossypol and increase tolerance to gossypol | 448 bp | Midgut | Arabidopsis RTM gene | pBI121 | Cotton | Larval growth retarded, insects displayed reduced feeding, down-regulation of target gene transcripts | – | Mao et al. (2011) | ||
| HaHR-3 | Molt-regulating transcription factor | >300 bp | Epidermis, midgut and fatbodies | – | pCAMBIA 2300 | Tobacco | Incomplete molting, reduction in larval weight, with significant down-regulation of target gene | Predicted similar effect due to conserve gene | Xiong et al. (2013) | ||
| 20-Hydroxyecdysone | Role in molting and metmorphosis | 482 bp | Gut, fat bodies And ovary | Arabidopsis RTM gene | pBI121 | Tobacco | Reduction in larval molting, pupation and adult emergence rate with silencing of target gene | Similar effect on S. exigua due to sequence homology | Zhu et al. (2012) | ||
| Arginine kinase | Play role in role in cellular energymetabolism | 1068 bp | Midgut, head and integument | GUS linker fragment | pANDA35HK | Arabidopsis thaliana | Detrimental effects on larval growth and survival, decrease in target gene transcripts | – | Liu et al. (2015) | ||
| 1. Chitin synthase 2. Cytochrome P450 monooxygenase 3. V-ATPase |
1. Synthesis of chitin 2. Required for tolerance against gossypol 3. Role in cellular energy production |
19 bp | 1. Trachea and epidermis and midgut 2. Midgut 3. Whole insect body |
9 bp loop | pLD | Tobacco | Reduction in target gene transcript level and reduction in weight, growth and pupation rate | – | Jin et al. (2015) | ||
| Chitinase | Role in molting and metamorphosis | 250 bp | Molting fluid | Chalcone synthase (ChaS) gene intron | pMVR-hp | Tobacco and tomato | Detrimental effects on larval growth and survival, decrease in larval and pupal weight, pupation and adult emergence with significant reduction in target gene transcript level | – | Mamta et al. (2016) | ||
| Diabrotica virgifera virgifera LeConte | VATPase -A | Role in cellular energy production | 246 bp | Whole insect body | – | pMON 94805 | Maize | Reduced feeding and stunted larval growth with decrease in target gene transcripts | No off-target effects | Baum et al. (2007) | |
| Spodoptera exigua | 20-Hydroxyecdysone | Role in molting and metamorphosis | 482 bp | Gut, fat bodies and ovary | Arabidopsis RTM gene | pBI121 | Tobacco | Partial lethality S. exigua, affected larval molting, pupation and adult emergence | Predicted similar effect due to conserve gene | Zhu et al. (2012) | |
| Menduca sexta | MsCYP6B46 | Nicotine degradation | 312 bp | Midgut | – | pSOL8 | Tobacco | Decrease in target gene transcripts and no effect on larval weight and survival | – | Kumar et al. (2012) | |
| Hemiptera | Nilaparvata Lugens | 1. Hexose transportergene 2. Carboxypeptidase 3. Trypsin like serine protease |
1. Transport of glucose 2. And 3 hydrolysis of protein |
<600 bp | Midgut | – | pKANNIBAL | Rice | Depletion in the transcript level and no effect on larval survival | – | Zha et al. (2011) |
| 20-Hydroxyecdysone | Role in molting and metamorphosis | 360 bp | Gut, Fat bodies And ovary | pSK-int vector | pCanG-HA | Rice | Depletion in the transcript level, reduction in fecundity | – | Yu et al. (2014) | ||
| Myzus persicae | 1. Rack1 2. MpC002 |
1. Essential in f oraging and feedingof thepea aphid 2. Intracellular receptor |
309 bp, 710 bp | 1. All parts of aphid body 2. Salivary glands |
– | pJawohl8- RNAi | Arabidopsis thaliana | Adversely affected survival rate and decreased fecundity | – | Pitino et al. (2011) | |
| hunchback(hb) | Play role in insect axial patterning | 427 bp | Embryo | – | pUCCRNAi | Tobacco | Reduced Mphb mRNA level in the fed aphids and inhibited insect reproduction | – | Mao and Zeng (2014) | ||
| Serine proteases (SP) | Gutproteolytic digestion | 550 bp | Gut | – | pANDA35HK | Arabidopsis thaliana | Significant attenuation of MySP expression and with noticeable reduction in fecundity | No off-target effects | Bhatia et al. (2012) | ||
| Bemisia tabaci | v-ATPase | Encode for v-ATPase subunit A, role in cellular energy production | 189 bp | Whole insect body | Arabidopsis RTM gene | pBI101 | Tobacco | Depletion in the transcript level and affected insect survival | – | Thakur et al. (2014) | |
| Acetylcholinesterase (AChE) and ecdysone receptor (EcR) | 1. Required in signaling 2. Role in molting and metamorphosis |
400 bp | 1. Whole insect body 2. Gut, fat bodies and ovary |
– | pJIT163+pCam bia2300 | Tobacco | Silencing of targets gene in chimeric construct produced detrimental effect on survival (appox. 80% mortality) with decrease in target genes transcripts | – | Malik et al. (2016) |
Based upon the above mentioned studies, it is evident that RNAi technology has a tremendous potential for control of all types of insect pests. In addition, RNAi technology coupled with Bt or other technologies offers a great choice in controlling the insects pests, which are prone to develop resistance against insecticidal proteins. However, to establish the true potential of HI-RNAi for combat of insect pests, further development and refinement of this technology in large-scale field tests are required. Similarly, like any other pest control strategy to be successful, the potential risks associated with RNAi technology need to be evaluated.
Conclusion and future directions
Recent studies have shown the potential of RNAi for the management of insect pests. RNAi involves the basic conserved mechanism and this feature makes it a suitable approach for control of all types of insect pests. However, the use of RNAi approach for control of pests at field level is still in its infancy and many road blocks need to be removed before establishing it as a viable insect pest control strategy. The continuous supply of dsRNA is required in sufficient amounts at the specific site for effective silencing of target genes due to the absence of silencing signal amplification system in insects. This problem could be overcome by identifying the vital genes, change in the expression, which will have detrimental effect on insect survival, and by engineering plants for the production of dsRNA molecules against such target genes. Many insect species including the economically important insect pests are getting sequenced and the availability of the whole genome sequences of these insects helps in better understanding of the RNAi machinery, identification of novel target genes and in overcoming the challenges faced during the application of RNAi approach as a pest control strategy. This new information will facilitate the future refinement of insect pest control methodologies based on RNAi and will continue to inspire discoveries of new strategies, which will provide new solutions to many of the existing and emerging problems related to the management of insect pests.
Acknowledgements
We are grateful to the Department of Biotechnology (Grant No. BT/AGR/TF/2006), New Delhi for generous support for RNAi work in the lab (to MVR). Mamta acknowledges the University Grants Commission, New Delhi for the senior research fellowship under the special assistance programme (SAP) of University Grants Commission (UGC). We also thank the UGC for SAP (DRS-III), Department of Science and Technology (DST), New Delhi for FIST (Level 2) programme and DU-DST PURSE (Phase II) Grant.
Compliance with ethical standards
Conflict of interest
The authors declare that they do not have conflict of interests.
References
- Allen ML, Walker WB. Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J Insect Physiol. 2012;58:391–396. doi: 10.1016/j.jinsphys.2011.12.014. [DOI] [PubMed] [Google Scholar]
- Andrade CE, Hunter WB. RNA interference—natural gene-based technology for highly specific pest control (HiSPeC) In: Abdurakhmonov IY, editor. RNA interference. Croatia: InTech; 2016. pp. 391–409. [Google Scholar]
- Araujo RN, Santos A, Pinto FS, Gontijo NF, Lehane MJ, Pereira MH. RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem Mol Biol. 2006;36:683–693. doi: 10.1016/j.ibmb.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo GM, Benoit JB, Michalkova V, Yang G, Roller L, Bohova J, Takácˇ P, Aksoy S. Analysis of lipolysis underlying lactation in the tsetse fly, Glossina morsitans. Insect Biochem Mol Biol. 2012;42:360–370. doi: 10.1016/j.ibmb.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardner R, Fletcher KE. Insect infestations and their effects on the growth and yield of field crops: a review. Bull Entomol Res. 1974;64:141–160. doi: 10.1017/S0007485300027061. [DOI] [Google Scholar]
- Batzri S, Korn ED. Single bilayer liposomes prepared without sonication. Biochim Biophys Acta. 1973;16:1015–1019. doi: 10.1016/0005-2736(73)90408-2. [DOI] [PubMed] [Google Scholar]
- Baum JA, Roberts JK. Progress towards RNAi-mediated insect pest management. Adv Insect Physiol. 2014;47:249–295. doi: 10.1016/B978-0-12-800197-4.00005-1. [DOI] [Google Scholar]
- Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- Belles X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu Rev Entomol. 2010;55:111–128. doi: 10.1146/annurev-ento-112408-085301. [DOI] [PubMed] [Google Scholar]
- Bhatia V, Bhattacharya R, Uniyal PL, Singh R, Niranjan RS. Host generated siRNAs attenuate expression of serine protease gene in Myzus persicae. PLoS One. 2012;7:e46343. doi: 10.1371/journal.pone.0046343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, et al. 3′ UTR seed matches, but not overall identities, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Boisson B, Jacques JC, Choumet V, Martin E, Xu J, Vernick K, Bourgouin C. Gene silencing in mosquito salivary glands by RNAi. FEBS Lett. 2006;580:1988–1992. doi: 10.1016/j.febslet.2006.02.069. [DOI] [PubMed] [Google Scholar]
- Bolognesi R, Ramaseshadri P, Anderson J, Bachman P, Clinton W, et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte) PLoS ONE. 2012;7:e47534. doi: 10.1371/journal.pone.0047534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher G, Scholten J, Klingler M. Parental RNAi in Tribolium (Coleoptera) Curr Biol. 2002;12:85–86. doi: 10.1016/S0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- Burand JP, Hunter WB. RNAi: future in insect management. J Invertebr Pathol. 2013;112:S68–S74. doi: 10.1016/j.jip.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Cappelle K, de Oliveira CF, Van Eynde B, Christiaens O, Smagghe G. The involvement of clathrin-mediated endocytosis and two Sid-1-like transmembrane proteins in double-stranded RNA uptake in the Colorado potato beetle midgut. Insect Mol Biol. 2016;25:315–323. doi: 10.1111/imb.12222. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christou P, Capell T, Kohli A, Gatehouse JA, Gatehouse AMR. Recent developments and future prospects in insect pest control in transgenic crops. Trends Plant Sci. 2006;11:302–308. doi: 10.1016/j.tplants.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Irelan JT, Schumacher M, Schmidhauser T, Selker EU, Macino G. Transgene silencing of the al-1 gene in vegetative cells of Neurosporais mediated by a cytoplasmic effectors and does not depend on DNA–DNA interactions or DNA methylation. EMBO J. 1996;15:3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Ganbaatar O, Cao B, Zhang Y, Bao D, Bao W, Wuriyanghan H. Knockdown of Mythimna separata chitinase genes via bacterial expression and oral delivery of RNAi effectors. BMC Biotechnol. 2017;17:9. doi: 10.1186/s12896-017-0328-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt JS, Belles X, Richards EH, Reynolds SE. Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: evidence from Manduca sexta and Blattella germanica. J Insect Physiol. 2013;59:171–178. doi: 10.1016/j.jinsphys.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Gordon KH, Waterhouse PM. RNAi for insect-proof plants. Nat Biotechnol. 2007;25:1231–1232. doi: 10.1038/nbt1107-1231. [DOI] [PubMed] [Google Scholar]
- Gu L, Knipple DC. Recent advances in RNA interference research in insects: implications for future insect pest management strategies. Crop Prot. 2013;45:36–40. doi: 10.1016/j.cropro.2012.10.004. [DOI] [Google Scholar]
- Hakim RS, Baldwin K, Smagghe G. Regulation of midgut growth, development, and metamorphosis. Annu Rev Entomol. 2010;55:593–608. doi: 10.1146/annurev-ento-112408-085450. [DOI] [PubMed] [Google Scholar]
- Hilder VA, Boulter D. Genetic engineering of crop plants for insect resistance—a critical review. Crop Prot. 1999;18:177–191. doi: 10.1016/S0261-2194(99)00028-9. [DOI] [Google Scholar]
- Howard KA, Rahbek UL, Liu X, Damgaard CK, Glud SZ, Andersen MØ, Hovgaard MB, Schmitz A, Nyengaard JR, Besenbacher F, Kjems J. RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system. Mol Ther. 2006;14:476–484. doi: 10.1016/j.ymthe.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Huang J, Pray C, Rozella S. Enhancing the crops to feed the poor. Nature. 2002;418:678–683. doi: 10.1038/nature01015. [DOI] [PubMed] [Google Scholar]
- Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol. 2010;56:227–235. doi: 10.1016/j.jinsphys.2009.10.004. [DOI] [PubMed] [Google Scholar]
- James C (2014) Global status of commercialized Biotech/GM crops. ISAAA Brief 49. ISAAA, Ithaca, NY
- Jin S, Singh ND, Li L, Zhang X, Daniell H. Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa armigera larval development and pupation. Plant Biotech J. 2015;13:435–446. doi: 10.1111/pbi.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joga MR, Zotti MJ, Smagghe G, Christiaens O. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front Physiol. 2016;7:553. doi: 10.3389/fphys.2016.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonathan GL, Jian JD. RNAi-based insecticidal crops: potential effects on nontarget species. Bioscience. 2013;63:657–666. doi: 10.1525/bio.2013.63.8.8. [DOI] [Google Scholar]
- Jose AM, Hunter CP. Transport of sequence-specific RNA interference information between cells. Annu Rev Genet. 2007;41:305–330. doi: 10.1146/annurev.genet.41.110306.130216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM, Smith JJ, Hunter CP. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc Natl Acad Sci USA. 2009;106:2283–2288. doi: 10.1073/pnas.0809760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann P, Schwirz J, Schmitt-Engel C, Bucher G. RNAi phenotypes are influenced by the genetic background of the injected strain. BMC Genom. 2013;14:5. doi: 10.1186/1471-2164-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knip M, Constantin ME, Thordal-Christensen H. Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet. 2014;10:e1004602. doi: 10.1371/journal.pgen.1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Biedenkopf D, Furch A, Weber L, Rossbach O, et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016;12:e1005901. doi: 10.1371/journal.ppat.1005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola VSR, Renuka P, Madhav MS, Mangrauthia SK. Key enzymes and proteins of crop insects as candidate for RNAi based gene silencing. Front Physiol. 2015;6:119. doi: 10.3389/fphys.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konakalla NC, Kaldis A, Berbati M, Masarapu H, Voloudakis AE. Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta. 2016;24:961–969. doi: 10.1007/s00425-016-2567-6. [DOI] [PubMed] [Google Scholar]
- Kumar M, Gupta GP, Rajam MV. Silencing of acetylcholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. J Insect Physiol. 2009;55:273–278. doi: 10.1016/j.jinsphys.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Kumar P, Pandit SS, Baldwin IT. Tobacco rattle virus vector: a rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS One. 2012;7:e31347. doi: 10.1371/journal.pone.0031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal OP. Field resistance of some tomato cultivars against the fruitworm, Heliothis armigera (Hubner). Tripoli. Bull Ent. 1985;26:46–47. [Google Scholar]
- Li H, Chougule NP, Bonning BC. Interaction of the Bacillus thuringiensis delta endotoxins Cry1Ac and Cry3Aa with the gut of the pea aphid, Acyrthosiphon pisum (Harris) J Invertebr Pathol. 2011;107:69–78. doi: 10.1016/j.jip.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Li H, Jiang W, Zhang Z, Xing Y, Li F. Transcriptome analysis and screening for potential target genes for RNAi-mediated pest control of the beet armyworm, Spodoptera exigua. PLoS One. 2013;8:e65931. doi: 10.1371/journal.pone.0065931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Guan R, Guo H, Miao X. New insights into an RNAi approach for plant defence against piercing-sucking and stem-borer insect pests. Plant Cell Environ. 2015;38:2277–2285. doi: 10.1111/pce.12546. [DOI] [PubMed] [Google Scholar]
- Lipardi C, Paterson BM. Identification of an RNA-dependent RNA polymerase in Drosophila involved in RNAi and transposon suppression. Proc Natl Acad Sci USA. 2009;106:15645–15650. doi: 10.1073/pnas.0904984106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu S, Ding Z, Zhang C, Yang B, Liu Z. Gene knockdown by introthoracic injection of double-stranded RNA in the brown planthoer, Nilaparvata lugens. Insect Biochem Mol Biol. 2010;40:666–667. doi: 10.1016/j.ibmb.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Liu J, Smagghe G, Swevers L. Transcriptional response of BmToll9-1 and RNAi machinery genes to exogenous dsRNA in the midgut of Bombyx mori. J Insect Physiol. 2013;59:646–665. doi: 10.1016/j.jinsphys.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Liu F, Wang XD, Zhao YY, Li YJ, Liu YC, Sun J. Silencing the HaAK gene by transgenic plant-mediated RNAi impairs larval growth of Helicoverpa armigera. Int J Biol Sci. 2015;11:67–74. doi: 10.7150/ijbs.10468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomazzo E, Hussmann GP, Wolfe BB, Yasuda RP, Perry DC, Kellar KJ. Effects of chronic nicotine on heteromeric neuronal nicotinic receptors in rat primary cultured neurons. J Neurochem. 2011;119:153–164. doi: 10.1111/j.1471-4159.2011.07408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HJ, Raza A, Amin I, Scheffler JA, Scheffler BE, Brown JK, Mansoor S. RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci Rep. 2016;6:38469. doi: 10.1038/srep38469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamta, Reddy KRK, Rajam MV. Targeting chitinase gene of Helicoverpa armigera by host-induced RNA interference confers insect resistance in tobacco and tomato. Plant Mol Biol. 2016;90:281–292. doi: 10.1007/s11103-015-0414-y. [DOI] [PubMed] [Google Scholar]
- Mao J, Zeng F. Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae. Transgenic Res. 2014;23:145–152. doi: 10.1007/s11248-013-9739-y. [DOI] [PubMed] [Google Scholar]
- Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen X. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- Mao YB, Tao XY, Xue XY, Wang LJ, Chen XY. Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Res. 2011;20:665–673. doi: 10.1007/s11248-010-9450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YB, Xue XY, Tao XY, Yang CQ, Wang LJ, Chen XY. Cysteine protease enhances plant-mediated bollworm RNA interference. Plant Mol Biol. 2013;83:119–129. doi: 10.1007/s11103-013-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehto DN, Singh KM, Singh RN. Incidence of insect pests in chickpea, Cicer arietinum L. Indian J Ent. 1985;47:117–136. [Google Scholar]
- Miguel SK, Scott JG. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag Sci. 2016;72:801–809. doi: 10.1002/ps.4056. [DOI] [PubMed] [Google Scholar]
- Miller SC, Brown SJ, Tomoyasu Y. Larval RNAi in Drosophila? Dev Genes Evol. 2008;218:505–510. doi: 10.1007/s00427-008-0238-8. [DOI] [PubMed] [Google Scholar]
- Miller SC, Miyata K, Brown SJ, Tomoyasu Y. Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS ONE. 2012;7:e47431. doi: 10.1371/journal.pone.0047431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous gene in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navale PM, Maligeppagol M, Mahadeva Swamy HM, Ramasamy A, Sowmya HD, Krishna V, Prasad Babu K, Kumbar BM. Plant mediated RNAi: a new line of defense against insect pests. Int J Biotechnol Appl. 2014;6:163–168. [Google Scholar]
- Nellen W, Lichtenstein C. What makes an mRNA antisenseitive? Trends Biol Sci. 1993;18:419–423. doi: 10.1016/0968-0004(93)90137-C. [DOI] [PubMed] [Google Scholar]
- Oerke EC. Crop losses to pests. J Agric Sci. 2006;144:31–43. doi: 10.1017/S0021859605005708. [DOI] [Google Scholar]
- Orii H, Mochii M, Watanabe K. A simple ‘‘soaking method’’ for RNA interference in the planarian Dugesia japonica. Dev Genes Evol. 2003;213:138–141. doi: 10.1007/s00427-003-0310-3. [DOI] [PubMed] [Google Scholar]
- Palli SR. RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Curr Opin Insect Sci. 2014;6:1–8. doi: 10.1016/j.cois.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Ni JQ, Perkins L. In vivo RNAi: today and tomorrow. Cold Spring Harb Perspect Biol. 2010;2:a003640. doi: 10.1101/cshperspect.a003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA. Silencing of aphid genes by dsRNA feeding from plants. PLoS One. 2011;6:e25709. doi: 10.1371/journal.pone.0025709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice K, Pertry I, Christiaens O, Bauters L, Bailey A, Niblett C, Ghislain M, Gheysen G, Smagghe G. Transcriptome analysis and systemic RNAi response in the African sweet potato weevil (Cylas puncticollis, Coleoptera, Brentidae) PLoS ONE. 2015;10:e0115336. doi: 10.1371/journal.pone.0115336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DR, Gatehouse JA. RNAi-mediated crop protection against insects. Trends Biotechnol. 2008;26:393–400. doi: 10.1016/j.tibtech.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Rinkevich FD, Scott JG. Limitations of RNAi of α6 nicotinic acetylcholine receptor subunits for assessing the in vivo sensitivity to spinosad. Insect Sci. 2013;20:101–108. doi: 10.1111/j.1744-7917.2012.01523.x. [DOI] [PubMed] [Google Scholar]
- Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- Roush DK, McKenzie JA. Ecological genetics of insecticide and acaricide resistance. Annu Rev Entomol. 1987;32:361–380. doi: 10.1146/annurev.en.32.010187.002045. [DOI] [PubMed] [Google Scholar]
- Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncb1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurabh S, Vidyarthi AS, Prasad D. RNA interference: concept to reality in crop improvement. Planta. 2014;239:543–564. doi: 10.1007/s00425-013-2019-5. [DOI] [PubMed] [Google Scholar]
- Shakesby AJ, Wallace IS, Isaacs HV, Pritchard J, Roberts DM, Douglas AE. A water-specific aquaporin involved in aphid osmoregulation. Insect Biochem Mol Biol. 2009;39:1–10. doi: 10.1016/j.ibmb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Sharma DK, Sharma T. Biotechnological approaches for biodiversity conservation. Indian J Sci Res. 2013;4:183–186. [Google Scholar]
- Sharma HC, Crouch JH, Sharma KK, Seetharama N, Hash CT. Applications of biotechnology for crop improvement: prospects and constraints. Plant Sci. 2002;63:381–395. doi: 10.1016/S0168-9452(02)00133-4. [DOI] [Google Scholar]
- Shukla JN, Kalsi M, Sethi A, Narva KE, Fishilevich E, Singh S, Mogilicherla K, Palli SR. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 2016;13:656–669. doi: 10.1080/15476286.2016.1191728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenersen J. Chemical pesticides: modes of action and toxicology. Boca Raton: CRC Press; 2004. [Google Scholar]
- Stevens J, Dunse K, Fox J, Evans S, Anderson M. Biotechnological approaches for the control of insect pests in crop plants. In: Soundararajan RP, editor. Pesticides-advances in chemical and botanical pesticides. Croatia: InTech; 2012. [Google Scholar]
- Tabara H, Grishok A, Mello CC. RNAi in C. elegans soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- Tabashnik BE, Brévault T, Carrière Y. Insect resistance to Bt crops: lessons from the first billion acres. Nat Biotechnol. 2013;31:510–521. doi: 10.1038/nbt.2597. [DOI] [PubMed] [Google Scholar]
- Taning CNT, Christiaens O, Berkvens N, Casteels H, Maes M, Smagghe G. Oral RNAi to control Drosophila suzukii: laboratory testing against larval and adult stages. J Pest Sci. 2016;89:803–814. doi: 10.1007/s10340-016-0736-9. [DOI] [Google Scholar]
- Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, et al. RNA interference in lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231–245. doi: 10.1016/j.jinsphys.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Thakur N, Upadhyay SK, Verma PC, Chandrashekar K, Tuli R, Singh PK. Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase A gene. PLoS ONE. 2014;9:e87235. doi: 10.1371/journal.pone.0087235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur N, Mundey JK, Upadhyay SK. RNAi-implications in entomological research and pest control. In: Abdurakhmonov IY, editor. RNA interference. Croatia: InTech; 2016. [Google Scholar]
- Tian H, Peng H, Yao Q, Chen H, Xie Q, Tang B, Zhang W. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS ONE. 2009;4:e6225. doi: 10.1371/journal.pone.0006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:85. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tomoyasu Y, Miller SC, Shuichiro T, Schomeier M, Grossmann D, Bucher G. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008 doi: 10.1186/gb-2008-9-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich J, Dao VA, Majumdar U, Schmitt-Engel C, Schwirz J, et al. Large scale RNAi screen in Tribolium reveals novel target genes for pest control and the proteasome as prime target. BMC Genom. 2015;16:674. doi: 10.1186/s12864-015-1880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, Rämet M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- Vinokurov KS, Elpidina EN, Oppert B, Prabhakar S, Zhuzhikov DP, Dunaevsky YE, Belozersky MA. Diversity of digestive proteinases in Tenebrio molitor (Coleoptera: Tenebrionidae) larvae. Comp Biochem Physiol. 2006;145:126–137. doi: 10.1016/j.cbpb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Walshe DP, Lehane SM, Lehane MJ, Haines LR. Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol Biol. 2009;18:11–19. doi: 10.1111/j.1365-2583.2008.00839.x. [DOI] [PubMed] [Google Scholar]
- Whyard S, Singh AD, Wong S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem Mol Biol. 2009;39:824–832. doi: 10.1016/j.ibmb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- Wynant N, Santos D, Verdonck R, Spit J, Van Wielendaele P, Vanden Broeck J. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol. 2014;46:1–8. doi: 10.1016/j.ibmb.2013.12.008. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Zeng H, Zhang Y, Xu D, Qiu D. Silencing the HaHR3 gene by transgenic plant-mediated RNAi to disrupt Helicoverpa armigera development. Int J Biol Sci. 2013;9:370–381. doi: 10.7150/ijbs.5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang XF, Chen P, Liu FT, Zheng SC, Ye H, Mo MH. RNA interference in moths: mechanisms, applications, and progress. Genes. 2016;7:88. doi: 10.3390/genes7100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue XY, Mao YB, Tao XY, Huang YP, Chen XY. New approaches to agricultural insect pest control based on RNA interference. Adv Insect Physiol. 2012;42:73–117. doi: 10.1016/B978-0-12-387680-5.00003-3. [DOI] [Google Scholar]
- Yencho GC, Cohen MB, Byrne PF. Applications of tagging and mapping insect resistance loci in plants. Annu Rev Entomol. 2000;45:393–422. doi: 10.1146/annurev.ento.45.1.393. [DOI] [PubMed] [Google Scholar]
- Yogindran S, Rajam MV, et al. RNAi for crop improvement. In: Bahadur Bir, et al., editors. Plant biotechnology: volume II: plant genomics and biotechnology. New Delhi: Springer; 2015. pp. 623–637. [Google Scholar]
- Yu R, Xu X, Liang Y, Tian H, Pan Z, Jin S, Wang N, Zhang W. The insect ecdysone receptor is a good potential target for RNAi-based pest control. Int J Biol Sci. 2014;10:1171–1180. doi: 10.7150/ijbs.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Christiaens O, Liu J, Niu J, Caelle K, Caccia S, Huvenne H, Smagghe G. Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci. 2013;20:4–14. doi: 10.1111/j.1744-7917.2012.01534.x. [DOI] [PubMed] [Google Scholar]
- Zha WJ, Peng XX, Chen RZ, Du B, Zhu LL, He GC. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the Hemipteran insect Nilaparvata lugens. Plos One. 2011;6:e20504. doi: 10.1371/journal.pone.0020504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang J, Zhu KY. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae) Insect Mol Biol. 2010;19:683–693. doi: 10.1111/j.1365-2583.2010.01029.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Li HC, Miao XX. Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci. 2013;20:15–30. doi: 10.1111/j.1744-7917.2012.01513.x. [DOI] [PubMed] [Google Scholar]
- Zhu JQ, Liu S, Ma Y, Zhang JQ, Qi HS, Wei ZJ, Yao Q, Zhang WQ, Li S. Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect associated gene EcR. PLoS ONE. 2012;7:e38572. doi: 10.1371/journal.pone.0038572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotti MJ, Smagghe G. RNAi technology for insect management and protection of beneficial insects from diseases: lessons, challenges and risk assessments. Neotrop Entomol. 2015;44:197–213. doi: 10.1007/s13744-015-0291-8. [DOI] [PubMed] [Google Scholar]