Abstract
Loss of the microvascular (MV) network results in tissue ischemia, loss of tissue function, and is a hallmark of chronic diseases. The incorporation of a functional vascular network with that of the host remains a challenge to utilizing engineered tissues in clinically relevant therapies. We showed that vascular-bed-specific endothelial cells (ECs) exhibit differing angiogenic capacities, with kidney microvascular endothelial cells (MVECs) being the most deficient, and sought to explore the underlying mechanism. Constitutive activation of the phosphatase PTEN in kidney MVECs resulted in impaired PI3K/AKT activity in response to vascular endothelial growth factor (VEGF). Suppression of PTEN in vivo resulted in microvascular regeneration, but was insufficient to improve tissue function. Promoter analysis of the differentially regulated genes in KMVECs suggests that the transcription factor FOXO1 is highly active and RNAseq analysis revealed that hyperactive FOXO1 inhibits VEGF-Notch-dependent tip-cell formation by direct and indirect inhibition of DLL4 expression in response to VEGF. Inhibition of FOXO1 enhanced angiogenesis in human bio-engineered capillaries, and resulted in microvascular regeneration and improved function in mouse models of injury-repair.
Keywords: Vascular regeneration, VEGF, angiogenesis, FOXO1, microfluidics, vascular rarefaction
Graphical abstract
Introduction
Angiogenesis, the process of creating new blood vessels from existing vessels, is vital in development, homeostasis, physiology, and in pathological responses to tissue damage. Proper tissue function requires a cell to be located within 150-200 μm of the nearest capillary [1] and it is without surprise therefore that most tissue engineering approaches rely on the presence of a functional vascular network [2-4]. The use of biomaterials in tissue engineering have focused on two major strategies: as a tool to control and prolong the release of angiogenic factors [5-7], or alternatively as a scaffold to enhance the growth of vascular cells supplied [3, 4, 8, 9].
The role of angiogenic factors and their use to vascularize engineered tissue constructs [10] as well as in vivo limb ischemia [11] have been well studied. However, their local regulation in specific microvascular beds of vital organs is less well understood. Angiogenesis occurs when an endothelial cell (EC) receives angiogenic cues that induce changes in its behavior resulting in its invasion and migration into the perivascular space to initiate blood vessel sprouting [12]. This highly coordinated and transient process involves the competition of neighboring ‘tip’ and ‘stalk’ cells that are morphologically and functionally distinct [13]. The leading ‘tip’ cell induces a neighboring EC to acquire tubular and supportive properties that enables elongation of the newly formed branch, thereby creating a new vessel. Studies in zebrafish intersegmental vessels [14, 15] and mouse retinal angiogenesis [16] show that the spatio-temporal regulation of DLL4 is instrumental in tip cell formation and angiogenesis.
The most well studied factor in therapeutic angiogenesis is vascular endothelial growth factor (VEGF) and biomaterials, such as hydrogels or silk scaffolds, have been used to successfully create a localized and sustained delivery of VEGF in vivo to enhance angiogenesis [6, 11, 17]. In the kidney, VEGF is highly expressed and tightly regulated in health as well as in response to injury [18, 19]. In healthy kidneys, high expression of VEGF contributes to the maintenance of endothelial cell fenestration in both glomerular and peritubular capillaries [20]. Following acute injury, however, overexpression of VEGF promotes deleterious effects on the renal microvasculature including vascular leak and inflammation with a limited effect on the formation of new blood vessels [21, 22]. Such observation hint that the kidney vasculature may have developed a limited angiogenic capacity as a protective mechanism to avoid excessive vascular leak and unnecessary angiogenesis. In fact, a reduction in VEGF signaling ameliorates renal function following injury [23]. Activation of VEGF receptor 2 (VEGFR2, FLK1, KDR), can initiate activation of several intracellular signaling cascades, including ERK/MAPK and PI3K/AKT, resulting in the transcriptional activation of the Notch ligand, Delta-like 4 (DLL4), a marker of tip-cell fate. Following the upregulation of the ligand DLL4 in response to VEGF signaling, DLL4 activates Notch signaling in the adjacent stalk cells resulting in the lateral inhibition of tip cell fates [13, 16]. Disruption of either the ERK/MAPK or the PI3K/AKT pathway impedes on angiogenesis [24], however little is known about how these two signaling pathways affect tip and stalk cell formation in a tissue-specific context. Recent work has shown that ERK is activated by VEGF in sprouting zebrafish intersegmental vessels and is required for angiogenesis [14]. Inhibition of ERK signaling resulted in marked reduction in DLL4 expression [14], as previously observed in studies of arterio-venous specification. During embryonic development, VEGF-dependent DLL4 expression in arteries is regulated by an enhancer region in the third intron, where binding of transcriptional regulators including ETS factor, and RBPJk have been shown to induce or maintain, respectively, the expression of DLL4. Studies using HUVECs suggest that VEGF-dependent induction of endothelial gene expression is transient, and can be mapped by H3K27Ac and EP300 binding to enhancer and promoter regions [25]. Whether E26 transformation-specific (ETS), EP300 or other transcriptional regulators are required for the induction of tip cell enriched genes, such as DLL4 is unclear.
Studies in the mouse indicate that the FOX-ETS transcription factor binding motif is enriched in endothelial specific enhancers and is synergistically activated by Forkhead box (FOX) and ETS families of transcription factors [26]. There are multiple ETS family members, but of these ETS-1 and ERG are highly expressed in the vasculature and have been shown to be regulated by VEGF signaling [27, 28]. The FOXO family of transcription factors has been implicated in metabolism, proliferation and longevity in endothelial cells [29, 30]. FOXO transcription factors are inhibited by AKT mediated phosphorylation of FOXOs leading to their nuclear exclusion and inactivity [31]. Of the FOXO class, FOXO1 mutation alone results in vascular defects early during development, which can be recapitulated by endothelial specific deletion of FOXO1 [32] suggesting an important role for FOXO1 in ECs. In this instance, an increase in EC proliferation was observed that has been shown to interfere with coordinated sprouting. However, examination of the mouse post-natal retina revealed differential localization of FOXO1 in the angiogenic front versus the plexus which suggest that coordinated angiogenesis requires tight regulation of FOXO1 activity [30]. In the adult, inducible knock out of the FOXO family improved neovascularization in ischemic tissues [33]. These observations indicate that FOXO1 plays a distinct role in adult tissues and that regulation of its activity is important during normal angiogenesis.
In disease states, the failure to adequately regenerate certain microvascular beds is closely linked to progressive organ failure. Such impaired vascular regeneration is implicated in highly prevalent diseases including stroke, liver cirrhosis, muscle atrophy, Alzheimer's disease as well as cardiac and kidney failure. The potential to use biomaterials to enhance vascularization of ischemic tissues [11, 34, 35], as well as for vascular tissue engineering applications [2, 4, 11] hold great promise. However, it should be noted that not all vascular beds respond similarly to angiogenic factors. In the following study, we dissected differences in the signaling pathways downstream of the VEGF receptor between distinct microvascular beds to identify the molecular underpinnings of impaired vascular regeneration. Using these novel insights, we demonstrate that microvascular regeneration can be successfully augmented to enhance functional organ recovery, and these strategies can be readily adapted in tissue engineering.
Materials and Methods
Cell isolation and assays
Mouse kidney MVECs were isolated from 6-8weeks C57BL/6 mice as described [36]. The single cell suspension was filtered with 40μm nylon mesh (BD Biosciences) to eliminate glomeruli and vessels and incubated with anti-CD31-PE (1:200, BD Biosciences) and anti-CD11b-APC (1:200, BD biosciences) for 5min on ice in FACS buffer (BD Biosciences) and sorted with FACS Aria (BD). Mouse kidney MVECs were expanded in RPMI supplemented with 20% FBS, 100 μg/ml endothelial cell growth supplement (ECGS, Lonza), 5U/ml heparin, 1× P&S for one passage and frozen down. Mouse AECs were cultured in Complete Mouse Endothelial Cell Medium (CellBiologics). Human kidney MVECs were purified from adult or fetal kidneys (IRB447773EA University of Washington) and New England Medical Center (Boston, MA) respectively using methods as described [37, 38] with modifications. Human kidney MVECs were characterized as a CD31 (BD 555446, 1:50) positive, PDGFRβ (Biolegend 18A1, 1:200) and CD45 (Biolegend H130, 1:400) negative population. Human lung, and heart MVECs were prepared in a similar manner and expanded in ECM (Sciencell). HUVECs, human and bovine AECs, were from Lonza. Mouse aortic endothelial cells (mAEC) were purchased from CellBiologics. To compare different cell types, cells between P2 and P7 were transferred to complete ECM medium (ScienCell) prior to experiment for at least 1 passage. Kidney MVECs were transfected at 50% confluency with 40 nM non-targeting control or 40 nM FOXO1 Silencer Select siRNA s5259 (Thermo Fisher Scientific) using RNAiMax (Thermo Fisher Scientific) and assayed after 72 hrs. Prior to stimulation, cells were serum starved in ECM basal medium supplemented with 0.5% FBS for 4 hours. Cells were treated with DMSO, 0.5μM FOXO1 inhibitor AS1842856 [39] (Selleckchem), or 3μM bpV [40] (EMD Millipore) for 1 hr before stimulated with 50ng/ml VEGF-A (R&D Systems), for durations stated.
Collagen invasion angiogenesis assays
Tissue explant experiments were performed as described [41, 42]. To stimulate angiogenesis, 50ng/ml VEGF-A was added and gels were imaged 4 days following explant. The 2-D collagen angiogenesis assay was performed as described with some modifications [43]. 40,000 endothelial cells were seeded on the collagen gel for 16 h in 96 well plates to allow for adhesion. Following a 4 hour serum starvation, the cells were stimulated with 50ng/ml VEGF-A (R&D Systems) in M199 supplemented with 10% FBS, 10μg/ml ascorbic acid (Sigma) and cultured for another 4 days. Cells were treated with 10μM LY294002 (Cell Signaling), or 1-10 bpV or 0.1-1 μM AS1842856. Cells were fixed with 3% glutaraldehyde (Sigma Aldrich) and stained with 0.1% toluidine blue O (Sigma) in 30% methanol for 30min. Each gel area (4×) was photographed and sprouting cells that invaded into collagen gel were counted. For lateral view imaging, gels were cut into 3mm slices and imaged using a microscope with 20× objective.
Protein analysis
100,000 ECs were seeded on 0.2% gelatin-coated 24-well plates and cultured overnight. Cells were serum starved for 4h and stimulated with 50ng/ml VEGF-A for 0, 5, or 15min. Cells or snap frozen whole kidneys were lysed in RIPA buffer containing Halt Protease and Phoshatase Inhibitor Cocktail (Thermo Fisher Scientific) on ice. Following protein quantification, SDS PAGE and Western blotting was performed as described [44]. The antibodies used for western blotting were from Cell Signaling unless otherwise specified and used at the following concentrations p-VEGFR2 (Y1175) (1:1000), P-AKT (S473) (1:2000), P-ERK (T202/Y204) (1:1000), and P-JNK (1:1000). P-FAK (1:1000, Biosource), P-p38 (1:1000, Sigma-Aldrich), P-SRC (Y416) (1:500), P-PTEN (S380) (1:400), P-FOXO1 (1:300, Abcam). Primary antibodies were detected with peroxidase conjugated anti-rabbit or anti-mouse IgG (1:3000) and visualized with SuperSignal West Femto Substrate (Thermo Fischer Scientific). Antibodies used to detect the total protein were as follows: AKT (11E7) (1:500), ERK (1:1000), FAK (1:1000), SRC (32G6) (1:500), FOXO1 (1:500), PTEN (1:500). Blots were quantified using ImageJ. In other experiments P-ERK was detected in ECs cultured on glass coverslips, following fixation and permeabilization as described using the anti-P-ERK antibody (1:500). All washes and buffers were prepared using 1× TBS. Nuclear vs cytoplasmic P-ERK was quantified using Image J.
RT-PCR and Quantitative PCR
Total RNA was extracted from cells or whole tissue using RNeasy Plus Mini Kit (Qiagen). Purity was determined by A260 to A280. cDNA was synthesized using oligo(dT) and random primers (iScript Reverse Transcription Supermix, Biorad). Quantitative PCR was performed using QuantStudio™ 7 Flex Real-Time PCR System (Life Technologies) using the TaqMan® Gene Expression Assays (Life Technologies). The specific primer pairs used in Q-PCR are listed in Table S1 (Life Technologies).
Microarray, RNA sequencing and Bioinformatics analysis
To compare different types of human endothelial cells, three different primary cell cultures for each cell type at passage 3 in identical conditions were harvested in RLT buffer and RNA was isolated. Automated sample amplifications and Biotin labeling was carried out using the NuGEN Ovation RNA Amplification system V2 (Cat # 3100), Ovation WB reagent (Cat # 1300) and Encore Biotin module (Cat # 4200) (NuGEN Technologies) according to manufacturer's recommendations using an Arrayplex automated liquid handler (Beckman Coulter). 2 μg of biotin labeled sscDNA probe were hybridized to Affymetrix GeneChip HT_MG-430_PM plate arrays with modified conditions as described in Allaire et al [45]. Washing and staining of the hybridized arrays were completed as described in the GeneChip Expression analysis technical manual for HT plate arrays using the Genechip® Array Station (Affymetrix, Santa Clara, CA) with modifications as described [46]. The processed GeneChip® plate arrays were scanned using GeneTitan scanner (Affymetrix, Santa Clara, CA). Affymetrix scans were subject to quality control (QC) measures. These tests included a visual inspection of the distribution of raw signal intensities and an assessment of RNA degradation, relative log expression (RLE), and normalized unscaled standard error (NUSE). All sample scans passed these QC metrics. CEL files were subjected to GC-content-based Robust Multi-array Average (GCRMA) normalization [47, 48]. Expression levels were log (base 2) transformed. All calculations and analyses were carried out using R and Bioconductor computational tools (ISBN: ISBN-10: 0-387-25146-4) Analyses were applied to discover genes that were differentially expressed (DEGs) between cell types of interest. To identify differentially expressed genes between groups of samples, we applied the linear modeling approach (Bioconductor limma package) to fit gene expression levels (log2 transformed) according to the defined groups of samples and Bayesian posterior error analysis as implemented by Smyth (DOI: 10.2202/1544-6115.1027). Genes that exhibited a log-odds score (lods) greater than zero, absolute fold change greater than 1.5, and average normalized intensity greater than four were considered significantly different. Pathway analysis was performed using Ingenuity Pathway Analysis tool (QIAGEN). To analyze for transcription factor activity, Whole Genome RVista was used (genome.lbl.org). The genomic regions surrounding Intron 3 of DLL4 were assessed for the presence of Evolutionary Conserved Regions (ECRs) using ECR Browser (ecrbrowser.dcode.org) and MuLAN (mulan.dcode.org) was used to identify conserved transcription factor binding sites using human as the base genome. MuLAN uses a global alignment tool. To detect FOXO1 regulated genes during angiogenesis, endothelial cells were plated for 2 days and serum starved for 4 hours prior to treatment with 0.5 μM AS1842856 [39] (Selleckchem) for 1 hour followed by a 30min treatment of VEGF-A. RNA was purified as described above from 4 separate endothelial cultures from different donors. RNA content was determined using next-generation sequencing (NGS) on the Illumina HiSeq 2000 platform producing 50bp paired-end reads. Following mapping of the reads and quality control evaluation, analysis was subjected to the enrichment test for gene ontology and pathway analysis using the Ingenuity Pathway Analysis tool (QIAGEN). Significant genes were selected based on p-values smaller than 0.05 and fold change greater than 2. For enrichment analysis of biological process ontology, differentially expressed genes were analyzed in DAVID 20 and processes were selected based on p-values smaller than 0.05.
Tissue analysis
Mouse tissues were collected and stained as previously described [49]. Primary antibodies used in this study were CD31 (1:200, BD Biosciences), FOXO1 (1:100, Cell Signaling), P-FOXO1 (1:100, Abcam), BrdU (1:200, Sigma Aldrich) and Ki67 (1:200, EMD Millipore), LTL (1:200, Vectorlabs), Phalloidin (1:50, Thermo Fisher Scientific), LAMA4 (1:200, Thermo Fisher Scientific) mounted in ProLong Gold Antifade Reagent with DAPI (Thermo Fisher Scientific). Images were captured by confocal (Intelligent Imaging Innovation or Zeiss) or standard fluorescent microscope (Nikon) as previously described [44]. Vessel fluorescence was analyzed at 400× magnification in images captured from CD31-stained sections of 10 different fields of S3 segment from at least five animals. Based on fluorescence intensities ranging from 0-255, peritubular capillaries were distinguished from background by empirically determining threshold values that marked only blood vessels in specimens from kidneys of sham operated mice. The threshold was constant for all measurements [50]. Kidney function was evaluated by measuring the urinary albumin to creatinine ratio (Albuwell) as described [51]. Confocal image stacks were analyzed using a Matlab (Mathworks) script to define the volume of the nucleus by thresholding the image on the DAPI channel. The CD31 fluorescence of the voxels immediately adjacent to the nucleus were averaged and compared to a threshold to sort the cells into endothelial positive or negative groups, and the FOXO fluorescence of the voxels inside the nuclei were averaged and compared to a threshold to sort the cells into FOXO positive and negative groups.
Microfluidic device set-up and assays
Nortis microfluidics platform (Nortis, Seattle, WA) was used to establish capillary beds in vitro as described [52], with the following modifications. The supporting collagen matrix was prepared at 2.5mg/ml (Life Technologies) on ice with 50ng/ml VEGF-A (R&D Systems). The ice-cold collagen mixture was injected into the Nortis devices using a 1ml syringe (Becton Dickinson), incubated on ice for 30min, RT for 30min, and finally at 37°C for 3h to allow for gelation. The 120μm fibers were pulled from the devices to create channels for cell seeding. The chips were perfused with ECM (ScienCell) at a continuous flow rate of 0.5ul/ml over night. To seed ECs, human MVECs were suspended at 7×106 cells/ml and injected into the channel with a microliter syringe (Hamilton) through an injection port in each device. The platforms were incubated statically for 1h to allow for cell attachment before flow was resumed. The chips were perfused at 0.5μl/ml and the culture media was supplemented with 50 ng/ml VEGF-A, 3μM bpV or 0.5 μM FOXO1 inhibitor. The medium was changed twice a day for 4 days. Devices were fixed with 4% PFA for 15 minutes using a multi-syringe pump (Braintree Scientific), washed in 1× TBS for 1h, blocked (1× Superblock (Cell Signaling), 5% FBS, 1% BSA, and 0.1% Triton X) for 1h. Primary antibodies were incubated overnight at 4°C in the same blocking buffer followed by washing in 1× TBS for 1h. Secondary antibodies were incubated for 1-1.5 hour followed by 2hrs washing with 1× TBS. The following primary antibodies were used: Phalloidin-rhodamine (1:500, Thermo Fisher Scientific), anti-CD31-Alexa594 (1:400, Biolegend), anti-VE-cadherin-Alexa488 (1:200, Biolegend). Devices were imaged using Zeiss confocal at 10× and 25× water immersion objectives. Sprout lengths and distance migrated were quantified using ImageJ.
Animal Models of Kidney Injury/Repair
Eight to ten weeks old male C57BL/6 mice, were anesthetized maintained with core temperature between 36.7 and 37.2°C and underwent clamping of the left renal pedicle for 23min, via a lateral flank incision. The kidney was confirmed to lose blood flow and go dusky. The right kidney was removed via a lateral flank incision, following surgical tying of the renal artery and vein (contralateral nephrectomy) as described [53]. Blood was collected from lateral tail vein at 1, 3, and 5 days after surgery for studies using bpV or days 3 and 7 for studies using AS1841674. Plasma creatinine and BUN were measured as described [51]. Mice were randomized to treatment and vehicle groups based on plasma creatinine levels 24h after surgery and received 2mg/kg bisperoxovanadium(HOpic) (EMD Millipore) via intraperitoneal injection (250μl/15mg body weight) and an equal volume of vehicle (PBS) to the control group. For studies using the FOXO1 inhibitor AS1842856 (Selleckchem), the compound was prepared in vehicle (0.5% methylcarboxycellulose, 0.1% Tween) and given at 100mg/kg by oral gavage 3 times a day from d3 to d7, to achieve a greater than IC50 coverage [39]. Mice were euthanized 5 days or 7 days after surgery. All the procedures followed protocols approved by Department of Comparative Medicine, University of Washington, or the IACUC at Biogen.
Chromatin IP and Luciferase Assays
ChIP was performed as described [54] using the Magna ChIP A/G Chromatin Immunoprecipitation Kit (EMD Millipore) with 1 μg of antibody: ERG (Santa Cruz), p300 (Santa Cruz), FOXO1 (Santa Cruz), and IgG (Sigma) was used as a negative control. Relative occupancy was calculated by determining the fold change of qPCR values for the specific antibody compared to IgG control. ChIP Forward Primer for DLL4 Intron 3: GTTTCCTGCGGGTTATTTTT and ChIP Reverse Primer for DLL4 Intron 3: CTTTCCAAAGGAGCGGAAT. Luciferase constructs containing the entire Intron 3 of the human DLL4 was synthesized (IDT technologies) and cloned into the pMCS minP-Tluc16-DD vector (Thermo Fisher Scientific) by adding SpeI and BsiWI adapters to the insert. RBPJk mutant construct was synthesized with mutations as described [54] and cloned as above. The human FOXO1 overexpression construct was purchased from OriGene. Luciferase experiments were performed as described using Bovine Aortic Endothelial Cells (Cell Applications Inc) cultured in ECM (Sciencell). 1 μg of each plasmid along with 0.01 μg of pCMV-Green Renilla Luc vector (Thermo Fisher Scientific) was transfected using Lipofectamine 2000 (Thermo Fisher Scientific) as per manufacturer's recommendations and luciferase assay was performed 18 hours later using the Dual Reporter Luciferase Assay (Promega). Luminescence was read using the Synergy Plate Reader (Biotek).
Statistics
All data are presented as mean ± SEM. Significance of difference between groups was evaluated by one-way ANOVA with Bonferroni post-hoc analysis or Student's t-test where appropriate (GraphPad Software). P values <0.05 considered to be significant and denoted with an *.
Results
The microvascular endothelium is resistant to angiogenesis and is characterized by deficient AKT and ERK responses to VEGFR-2 activation
We evaluated whether inherent resistance to form new capillaries exists in our internal organs, by performing an angiogenic sprouting assay on whole 1-2mm3 pieces of murine explanted organs embedded in collagen matrix in the presence or absence of VEGF-A (Fig 1A-B). Large vessels showed the greatest propensity to form sprouts and new capillaries whereas internal organs, including CNS, heart, lung, and trachea, showed a deficit. Importantly, the kidney did not form any new capillaries, despite viability of the endothelium within the explanted tissue. Moreover, large vessel explants (vein and aorta) showed marked responsiveness to exogenous VEGF (Fig 1A). However, these results can be challenging to interpret as it is difficult to control the cell types present in each organ chunk, surface area, as well as the endothelial and stromal cell densities. It is worth noting that vascular density in the healthy kidney is as high as any other organ, and pericyte density is estimated at 1 pericyte per endothelial cell [55]. To explore the significance of this observation to vascular regeneration in the setting of disease, murine kidneys were subjected to ischemia reperfusion injury (IRI) —a common injury resulting in rarefaction of the vasculature in humans [56]. In this controlled model of disease, injury leads to subsequent progressive loss of vascular density instead of complete restoration (Fig 1C, Fig S1A). By contrast, skin-wounding results in enhanced vascularity over the same time course [57, 58]. To understand why murine kidney capillaries failed to undergo angiogenesis, we FACS sorting to prepare cultures of kidney microvascular endothelial cells (KMVECs) (Fig S1B). Murine KMVECs expressed typical endothelial receptors and ligands (Fig S1C), yet in 2D collagen invasion assays, in which endothelial monolayers undergo sprouting angiogenesis into a collagen gel, KMVECs did not form sprouts even in the presence of VEGF-A (Fig 1D). In contrast, HUVECs, human cardiac MVECs or murine AECs had robust angiogenic responses in this assay (Fig 1D and S1H). To determine whether this finding was recapitulated in humans, we purified and expanded human KMVECs [37]. In the collagen invasion assay, human KMVECs similarly lacked angiogenic capacity (Fig 1E), even though human KMVECs expressed similar levels of FLT1, KDR, NRP1, and NRP2 in comparison to HUVECs (Fig S1D-G respectively). Although KMVECs expressed comparable levels of KDR (VEGFR2) to HUVECs (Fig S1E) it was possible that activation (phosphorylation) of VEGFR2 was deficient. However, we did observe VEGFR2 phosphorylation in murine KMVECs, albeit at a slightly lower level than murine AECs (Fig 1F). Despite this similarity in VEGFR2 activation, the subsequent activation of one of the major VEGFR2 signaling pathways, controlled by PI3 kinase and AKT was markedly reduced in mouse KMVECs (Fig 1F). Murine KMVECs were further evaluated for VEGFR2 responsiveness and activation of downstream signaling pathways ex vivo in comparison to HUVECs, which showed the greatest levels of angiogenesis in the 2D collagen sprouting assay (Fig 1D and S1H). Mouse KMVECs showed a slight reduction in VEGFR2 activation (Fig 1G), but activated AKT to a much lower level. In addition, the activation of ERK in mKMVECs was substantially abbreviated and attenuated (Fig 1G), whereas FAK (Fig 1G) and SRC (Fig S1I) activation were similar. To test the importance of AKT signaling and ERK signaling to sprouting angiogenesis, we developed a functioning 160 μm diameter capillary using HUVECs in Nortis microfluidic devices with luminal flow controlled by pumps (Fig 1H). The device matrix contained VEGF-A to promote angiogenesis and VEGF-A was further supplied in the medium. HUVECs, robustly formed new vessel sprouts with tip cells and long branching capillaries (Fig 1I). Inhibition of AKT signaling was established by the addition of LY294002, a PI3Kinase inhibitor, and the formation of new capillaries was significantly attenuated (Fig 1I), a finding replicated in the collagen invasion assay (Fig S1J-K). In addition, ERK inhibition, achieved using the inhibitor U0126, markedly reduced sprouting angiogenesis in the microfluidic devices, and notably, resulted in the presence of individual endothelial cells migrating from the vessel without forming new capillaries (Fig 1I). We hypothesized therefore that impaired angiogenic potential in KMVECs may be caused by endogenous disruption in AKT signaling or attenuated ERK signaling in response to VEGFR2.
Fig 1. Deficient angiogenesis from solid organ endothelium is associated with attenuated ERK and AKT signaling.
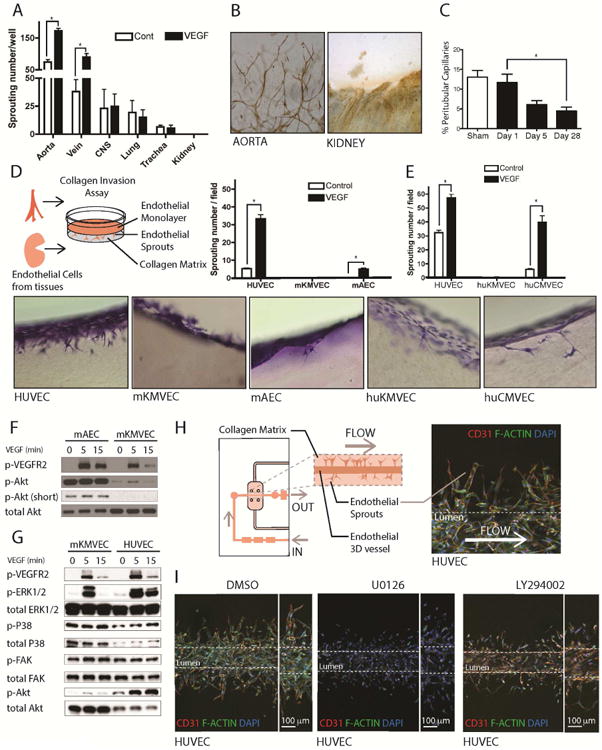
(A) Graph of angiogenic sprouts 72h after placing 1 mm3 pieces of internal organs in collagen gel in the presence or absence of 50ng/ml VEGF-A. (B) Light micrographs showing CD31-stained (brown) new vessels sprouting from aorta but not from kidney (C) Graph showing the % area of peritubular capillaries in the kidney following IRI. (D-E) Schema, toluidine blue stained thick sections and graphs showing angiogenesis in a gel invasion assay of mouse (D) or human (E) endothelial cells. Note MVECs from both mouse and human kidney showed very little capacity to form angiogenic sprouts and this was not enhanced by the addition of VEGF-A. (F) Blots showing activated (phosphorylated) VEGFR2, AKT in mAECs compared to m KMVECs. Note that KMVECs show very little activated AKT. (G) Bots showing activated VEGFR2, ERK, P38, FAK and AKT comparing mouse KMVECs and HUVECs. Note KMVECs have attenuated p-ERK1/2, and almost absent p-AKT in response to VEGF. (H) Schema and image of angiogenesis in human capillaries microfabricated in microfluidic devices. (I) Images of human capillaries generated from HUVECs under flow, 4 days after stimulating the vessels with VEGF-A in the surrounding gel and lumen to induce angiogenesis. Note widespread formation of stalks into the gel and the presence of tip cells with filopodia in the presence of vehicle (DMSO). However, the in the presence of the MEK inhibitor U0126, HUVEC cells migrate away from the vessel into the surrounding matrix, however, they are unable to form connections to the parent vessel or tip-and-stalk structures. In the presence of LY294002, which inhibits PI3K, new capillary formation is markedly attenuated with individual cells migrating out of the vessel wall similar to but less extensive than the effect of U0126. (n = 5 per group; P < 0.05)
Constitutively active PTEN suppresses AKT signaling and can be overcome to stimulate microvascular regeneration
The lack of AKT activation in KMVECs in response to angiogenic ligands appeared to be a finding that could explain why kidney and potentially other microvascular endothelia are predisposed to loss of the microvascular network following injury [33]. The activation of AKT is controlled by phosphoinositide-3 kinase (PI3 kinase) activity on the one hand, and by phosphatase activity on the other hand. Phosphatase and tensin homolog (PTEN) is a major endogenous phosphatase known to counteract PI3 kinase activity [59]. In response to VEGF-A mouse KMVECs retain high levels of the active (unphosphorylated) form of PTEN, consistent with PTEN forming a negative feedback loop on the AKT pathway (Fig 2A). We did not observe differences in other pathways, such as SRC kinase (Fig S1I). These signaling responses in mouse KMVECs were reproduced in human KMVECs (not shown). A small molecule inhibitor of PTEN known as bisperoxovanadium(HOpic), abbreviated to bpV, was applied to KMVECs permitting VEGF-A to activate AKT (Fig 2B). In the collagen invasion assay, bpV effectively restored angiogenic potential in mouse (Fig 2C) and human KMVECs (Fig 2D). To validate the capacity of bpV to enhance or restore angiogenic capacity in a functioning capillary, we generated human KMVEC capillaries with continuous flow using microfluidic devices made by Nortis, in which angiogenesis was stimulated by VEGF-A in the collagen matrix (Fig 2E). Human KMVECS in control conditions formed a complete capillary, and small numbers of KMVECs migrated into the matrix and a few formed small sprouts under the influence of VEGF-A. The KMVEC capillaries, under angiogenic stimulation, looked similar to HUVECs in the presence of the ERK inhibitor (Fig 1I). When bpV was perfused through the lumen, KMVECs could form large sprouts that are connected to the parent vessel (Fig 2E), confirming the capacity of PTEN inhibition to restore angiogenic capacity to human kidney capillaries. To test the significance of these observations in vivo, bpV was given to mice 1 day after IRI and continued to 5 or 7 days (Fig 2F-G) at concentrations shown to enhance AKT signaling 3-fold. These time points were chosen as the phase of organ regeneration after injury in this model (Fig 2G). BpV had no effect on vascularity in the healthy adult kidney (Fig 2H), and enhanced the phosphorylation of AKT as expected (Fig 2I). Although there was no difference in vascular density at Day 5 after injury, bpV protected against the loss of capillaries evaluated by CD31 density at Day 7, as well as stimulated greater proliferation of KMVECs (Fig 2H, J-K). At the same time a greater number of ECs not associated with laminin-a4 basal lamina was identified in the mice treated with bpV, consistent with an increase in tip cells (Fig 2L, Fig S2A). These bpV stimulated changes led to the increase in peritubular capillaries (PTCs) compared to vehicle (Fig 2M). Consistent with these observations, transcript levels for tip cell markers Apln, Vegfr2 and Dll4 were enhanced by bpV treatment, although not significant (Fig S2B). We did not observe changes in tubular injury in vehicle control versus bpV treated groups (Fig S2C), however, it is possible that a longer treatment period is required to observe differences.
Fig 2. High levels of active PTEN in MVECs can be overcome by the inhibitor bpV to restore angiogenesis and stimulate microvascular regeneration.
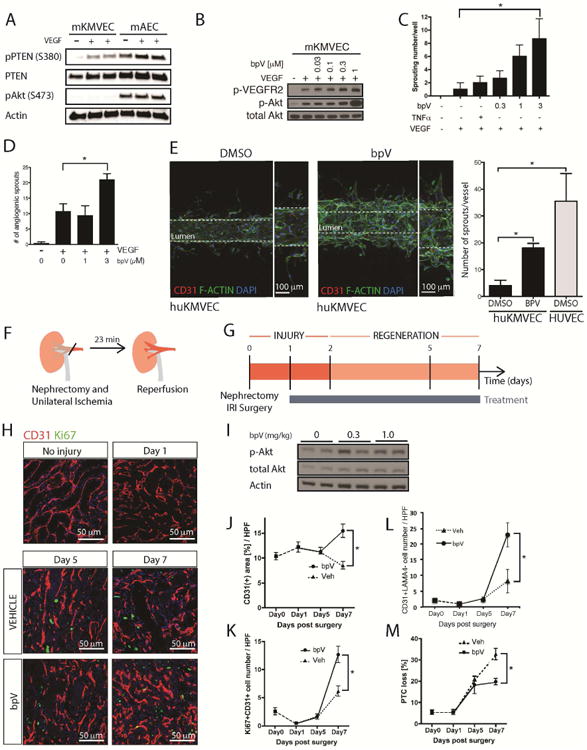
(A) Blots showing levels of total and inactive (phosphorylated) PTEN as well as active AKT. KMVECs have higher levels of active PTEN. (B) Blots showing the effect of concentrations of the PTEN inhibitor bpV on activate AKT in response to VEGF in mouse ECs. (C-D) bpV permits successful angiogenesis in the gel invasion assay of mouse (C) and human (D) MVECs from kidney. (E) In microfluidic devices under flow human kidney MVECs do not undergo sprouting angiogenesis in response to VEGF-A (left panel), rather individual cells migrate from the capillary, but are not connected to the lumen wall. By contrast (right) in the presence of bpV, successful angiogenesis occurs with sprouts that are connected to the parent vessel. In direct comparison bpV permits KMVECs to form new capillary sprouts at approximately half the level shown by HUVECs. (F-G) Schema and timecourse showing injury/repair caused by IRI in mouse kidney, the phase of regeneration and the duration of treatment with bpV. (H) Representative fluorescence images of kidneys after IRI, from mice treated with vehicle or bpV (I) Western blots of whole kidneys showing bpV effect on P-AKT (J-K) Effect of bpV on CD31 area (J) and proliferating ECs (K) in whole tissue sections (L) Effect of bpV on tip cell formation as defined by EC sprout without associated basal lamina detected by lamanina4 (M) Effect of bpV on total capillary loss. (n = 5-7 animals per group; P < 0.05).
Human endothelium with impaired angiogenesis is characterized by suppressed AKT activity leading to enhanced FOXO1 activity
To gain a deeper understanding of the differential programing of microvascular endothelium, we compared the steady-state transcriptome of large vessel and microvascular endothelium, including KMVECs. Lung MVECs were used as a comparison since they showed intermediate angiogenic capacity (Fig 1A). In identical culture conditions, we discovered a pattern of differentially expressed genes (DEGs), whereby HAECs and HUVECs showed the greatest difference from KMVECs, and lung MVECs the fewest differences (Fig S3A). Hierarchical clustering showed KMVECs to be highly enriched for a distinct set of genes, and lung MVECs were also partially enriched for these genes compared to large vessel ECs (Fig S3A). A distinct set of 188 genes were enriched specifically in KMVECs compared to AECs and HUVECs (Fig 3A), and when these genes were subjected to upstream analysis for impacted pathways, KMVECs were enriched for inhibition of the PI3 Kinase/AKT pathway (LY294002) and negatively enriched for VEGF signaling pathway (Fig 3B), consistent with our findings above. When the 188 enriched genes were evaluated for transcription factor binding sites in their promoters, they were found to be enriched for AP4 and FOXO factors (Fig 3C). Additional evaluation of transcription factor binding sites in the promoters of the VEGF regulated genes (Fig 3B) also identified FOXO factors as transcriptional regulators of those genes (Fig 3D). The four FOXO factors have conserved binding site consensus sequences, but of the FOXO factors, only mutation in FOXO1 results in a vascular phenotype [32]. Consistent with our observations that KMVECS have enhanced PTEN activity leading to a decrease in active AKT signaling, we hypothesized therefore that KMVECs had enhanced FOXO1 transcription factor activity, as AKT has previously been shown to inhibit FOXO1 function [60]. Using a high affinity inhibitor of FOXO1, AS1842856, which prevents FOXO1 binding to chromatin [39], we evaluated whether FOXO1 inhibition could restore angiogenesis. We generated human kidney capillaries using Nortis microfluidic devices (Fig 3G), with VEGF embedded in the matrix, as well as supplied in the media luminally. In addition, the lumen was perfused with a small molecule inhibitor of FOXO1 activity, AS1842856. After 4 days, the formation of new capillaries was evaluated (Fig 3E). Unlike HUVECs, KMVEC capillaries had little propensity to form new vessels, and in areas where this was attempted, thin capillaries or individual endothelial cells were detected outside of the lumen wall (Fig 3E DMSO control). By contrast, human kidney capillaries treated with the FOXO1 inhibitor showed robust formation of new sprouts with clear tip cell formation, and long branch lengths (Fig E). To further validate these observations, we generated capillaries using human KMVECs derived from adult hypertensive donors over 40 years of age who might be expected to have chronically diseased microvasculature. Similar to the healthy kidney endothelium, many endothelial cells migrated from the capillary without forming new vessels in the control group (Fig 3F). Importantly, older adult human KMVECs underwent some sprouting, but these sprouts showed attenuated branching, and formation of few filopodia at the angiogenic front (Fig 3F DMSO control, S3B). FOXO1 inhibition restored angiogenic capacity as well as tip cell identity (Fig 3F). bpV, in parallel experiments showed weaker effects on new vessel formation in these aged adult cells (Fig 3F). These results suggest that FOXO1 inhibition may allow KMVECs to respond better to VEGF-A induced signaling and to perhaps adopt tip and stalk cell identities, although more detailed time lapse analyses would be required to validate this hypothesis.
Fig 3. KMVECs are characterized by enhanced FOXO1 activity that can be inhibited to restore angiogenesis in functioning human kidney capillaries.
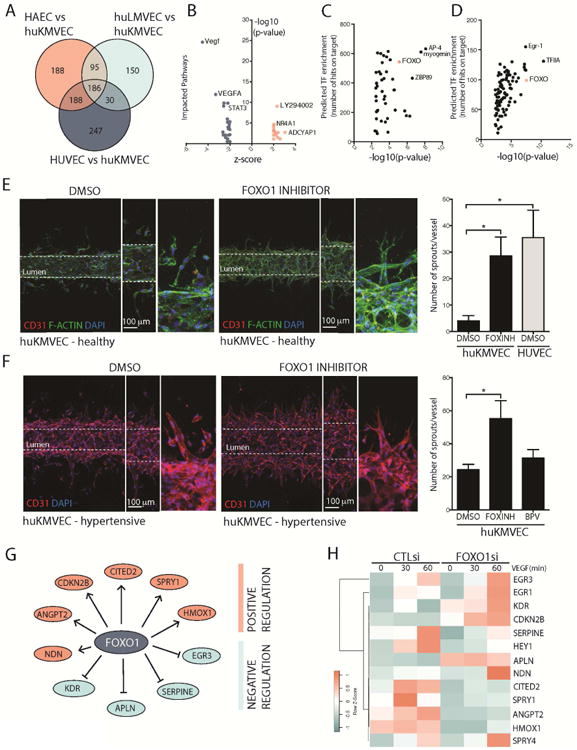
(A) Venn diagram showing overlapping and unique genes expressed by different types of endothelial cells. (B) Upstream analysis of the 186 genes enriched in KMVECs compared to HAECs showing genes consistent with impaired VEGF signaling as well as STAT3 signaling, and expression of genes consistent with blockade of PI3K/AKT by LY924002. (C-D) Transcription factor binding to promoters of DEGs unique to KMVECs (C) and unique to VEGF regulated genes identified in B (D) shows a large number of conserved FOXO1 binding sites at these promoters, thus pointing to enhanced FOXO1 activity (E-F) Images and results from microfluidic devices containing human kidney capillaries from young healthy kidneys (n=6 per group, 2 donors) (E) or aged hypertensive kidneys (n=5 per group, 1 donor) (F), stimulated to undergo sprouting angiogenesis with VEGF. In the presence of the FOXO1 inhibitor AS1842856 both healthy capillaries and aged capillaries successfully form new capillaries with tip-stalk structures. FOXO1 inhibition enables angiogenic capacity in kidney MVECs like that seen in HUVECs. Capillaries engineered from aged hypertensive kidneys had a higher tendency to form new vascular structures, although these did not show tip cell morphology (F, lower left images). FOXO1 inhibition enhanced new vessel formation with obvious tip cell morphology. Note bpV was less effective in aged capillary. (G) Pathway analysis of the DEGs in KMVECs showed genes predicted to be regulated by FOXO1 (H) Effect of siRNA against FOXO1 in KMVECs on potential FOXO1 target genes from G while under dynamic stimulation by VEGF-A by quantitative RT-PCR. Note CITED2 and SPRY1, identified as novel targets of FOXO1, show marked dynamic regulation by VEGF-A.
Inhibition of FOXO1 allows KMVEC to respond to VEGF-A induced signaling
Although the VEGF receptor, KDR, can be activated following VEGF-A treatment in KMVECs (Fig 1F-G), we have shown that activation of AKT is decreased, leading to enhanced FOXO1 activity. As a result, we observed a reduction in filopodia formation (Fig S3B) and impaired angiogenesis (Fig 3E-F) in KMVECs that could be rescued by inhibiting FOXO1 transcriptional activity. These observations collectively therefore were consistent with FOXO1 acting as a suppressor of angiogenesis in the kidney endothelium. We therefore asked how inhibition of FOXO1 could restore KMVECs' response to VEGF-A signaling, which is highly dynamic during angiogenesis. Through upstream pathway analysis, we found that FOXO1 can regulate a number of VEGF dependent genes such as KDR, APLN, and EGR3 (Fig 3G). RNA silencing of FOXO1 in KMVECs followed by dynamic VEGF-A stimulation for 30 or 60 minutes, confirmed activation of the VEGF-dependent genes found through upstream analysis, as well as CITED2, SPRY1, and EGR1 by quantitative PCR (Fig 3H, S3C). In addition, RNA silencing of FOXO1 in KMVECs prominently suppressed known FOXO1 targets, such as HMOX1, and ANGPT2 (Fig 3H). We were especially intrigued by FOXO1's regulation of SPRY1, as it is both regulated by VEGF-A signaling, and is an endogenous inhibitor of ERK signaling from tyrosine kinase receptors including KDR [61, 62]. Elevated SPRY1 expression in KMVECs (Fig 3H, S3C) may explain the attenuated activation of ERK signaling in response to VEGF-A induction (Fig 1G). One caveat of the single channel microfluidic system by Nortis, however, is the lack of a VEGF gradient, usually present in many models of angiogenesis in vivo. In order to evaluate the impact of FOXO1 mediated-suppression of the VEGF signaling pathway in KMVECs, we used a dual lumen microfluidic device under gravity driven flow [63]. This device provides a standing gradient of VEGF-A from one lumen across a matrix to the capillary, providing directional cues and allowing for more physiological angiogenesis. In these devices, significant migration of KMVECs occurred but there was little sprouting, and sprout length was short. FOXO1 inhibition increased sprouting (Fig 4A), and increased the length of new vessels formed (Fig 4A). These findings are consistent with FOXO1 having effects throughout the angiogenic process including the formation of robust vessels.
Fig 4. FOXO1 suppresses VEGF-A mediated nuclear ERK signaling to prevent activation of the tip cell marker DLL4.
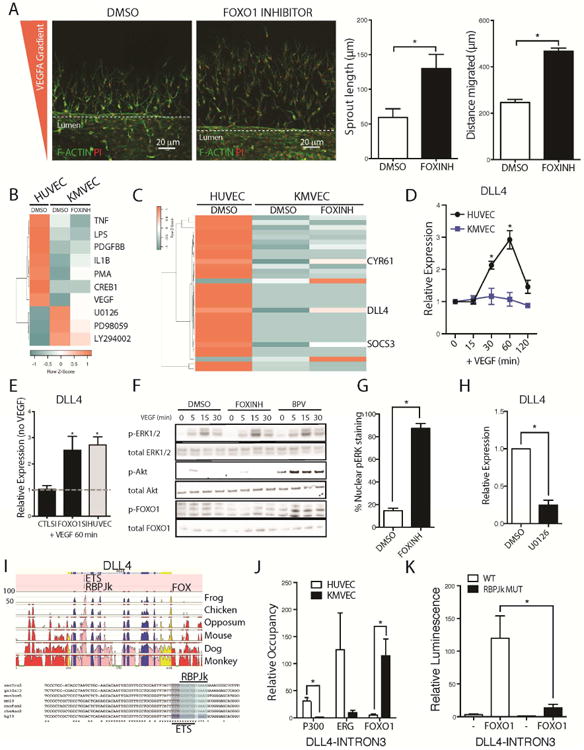
(A) Images from a dual channel microfluidic system with a constant lateralized VEGF gradient showing that inhibition of FOXO1 induces greater response to VEGF in KMVECs which results in both increased sprout length and increased distance migrated (B) Heatmap showing upstream pathways, detected by RNA sequencing and analyzed by IPA, hierarchically clustered that are enriched in HUVECs, and KMVECs following 1 hr treatment with FOXO1 inhibitor and 30 mins stimulation with VEGF-A (C) Heatmap showing hierarchical clustering of the genes most enriched in HUVECs and KMVECs following treatment with FOXO1 inhibitor and 30 mins after treatment with VEGF-A. DLL4 is most strongly de-repressed when FOXO1 was inhibited. Other genes include CYR61 and SOCS3 (D) Time course of DLL4 transcription in HUVECs and KMVECs in response to VEGF-A. KMVECs are unresponsive. (E) Knock down of FOXO1 in KMVECs restored activation of DLL4 expression in response to VEGF-A (F) Blots showing bpV enhances p-AKT and p-FOXO1 but neither inhibitor enhances total pERK in MVECs. (G) Quantification of pERK localization within KMVECs following stimulation by VEGF-A for 60 minutes in the presence and absence of FOXO1 inhibitor. pERK is predominantly cytoplasmic in KMVECs after VEGF-A, but in the presence of FOXO1 inhibitor it is predominantly nuclear (H) Q-PCR results showing in expression of DLL4 is reduced when MEK is inhibited. (I) Conserved putative transcription factor binding sites within the DLL4 locus, specifically focusing in on Intron 3 where both ETS and RBPJk sites are 100% conserved between many vertebrates. (J) ChIP of EP300 and ERG show low binding to the DLL4 intron 3 in KMVEC compared to HUVEC with the converse observed with FOXO1 occupancy at this locus. (K) Luciferase assay with a reporter construct containing the Intron 3 of DLL4 as well as the RBPJk mutant version. Overexpression of FOXO1 induces the wild type construct but not the RBPJk mutant (n = 3-6/group; * P < 0.05).
FOXO1 inhibits VEGF-A dependent induction of DLL4
To identify FOXO1-regulated factors, that control the initiation of angiogenesis, we sequenced RNA from human KMVECs and HUVECs 30 min after VEGF-A stimulation in the presence or absence of FOXO1 inhibition (GSE93590). Pathway analysis revealed that the VEGF signaling pathway was suppressed in KMVECs, and can be partially reversed by FOXO1 inhibition (Fig 4B). As predicted, MEK/ERK and PI3K/AKT signaling was suppressed in KMVECs, as demonstrated by pathways controlled by U0126 (MEK), PD98059 (MEK) and LY294002 (PI3K) (Fig 4B). We identified 35 genes that are regulated by FOXO1 in KMVECs. These FOXO1 target genes include SPRY1 and CITED2, which we had identified as FOXO1 effectors, but additionally include several LncRNAs whose functions are unclear (Fig S4A). When we ranked genes that are suppressed in KMVECs but not in HUVECs in response to VEGFA, which can be rescued by the inhibition of FOXO1, we found DLL4 to be most strongly de-repressed when FOXO1 was inhibited (Fig 4C). Other genes include CYR61 and SOCS3 (Fig 4C). Spatio-temporal regulation of the Notch ligand DLL4 is instrumental in tip cell formation and angiogenesis, and its transcriptional regulation has been shown to be downstream of VEGF-dependent MAPK signaling[14]. We therefore sought to investigate the role of FOXO1 in regulating DLL4 transcription, which is previously not known. Although levels of DLL4 expression is the same in HUVECs as in KMVECs (Fig S4B), VEGF-A rapidly and transiently induces expression of DLL4 only in HUVECs, but not in KMVECs (Fig 4D). As predicted, silencing of FOXO1 expression by siRNA was sufficient to restore DLL4 induction in response to VEGF-A in KMVECs, to levels seen in HUVECs (Fig 4E). However, it should be noted that although silencing of FOXO1 can result in induction of DLL4 expression in response to VEGF-A, the total amount of DLL4 is reduced (Fig S4C). This suggests that not only does FOXO1 repress induction of DLL4 upon VEGF-A stimulation, but FOXO1 could function to maintain DLL4 expression as well. We first focused on the induction of DLL4 by VEGF-A signaling pathways. As we identified SPRY1 as a gene target for FOXO1, we anticipated that FOXO1 inhibition would result in enhanced ERK signaling in response to VEGF-A, yet this was not apparent by whole cell blotting (Fig 4F). In single KMVEC cells however, although ERK was phosphorylated in response to VEGF-A, it did not enter the nucleus (Fig S4D). In the presence of FOXO1 inhibition pERK rapidly accumulated in the nucleus (Fig S4D, Fig 4G). Whether this is due to a reduction of SPRY1 levels when FOXO1 is inhibited is not clear, and further experiments need to be performed to test this hypothesis. We further observed that when HUVECs are treated with the MEK inhibitor U0126, the expression of DLL4 is significantly reduced (Fig 4H).
FOXO1 controls transcriptional complex association at the VEGF-dependent DLL4 enhancer in Intron 3
As phospho-ERK is thought to directly phosphorylate target nuclear factors including ETS transcription factors to drive the expression of DLL4 [14, 54], we hypothesize that inhibition of FOXO1 relieves the inhibition of pERK and allows for transcription factors such as ETS to regulate DLL4 expression. Wythe et al (2013) recently described a VEGF-dependent enhancer in Intron-3 of DLL4 (Fig 4I) [54]. This enhancer has binding sites for ETS factors, and the Notch transcriptional effector, RBPJk (Fig 4I). To determine the mechanism of how FOXO1 controls the transcriptional activity of DLL4, we looked at recruitment of EP300, the ETS factor ERG and FOXO1 following a 20 minute VEGF stimulation in both HUVECs and KMVECs using Chromatin immunoprecipitation (ChIP). Although FOXO1 has a conserved binding motif in the 3′ UTR region of DLL4, there is no conserved FOXO1 binding site at Intron-3 of DLL4 (Fig 4I). The 3rd intronic enhancer locus has binding sites for ETS1/ERG and RBPJk which positively regulate transcription. We observed that HUVECs showed enhanced recruitment of both ERG and EP300 at the DLL4-Intron3 enhancer, with very little FOXO1 occupancy, in response to VEGF stimulation (Fig 4J). KMVECs on the other hand, showed high FOXO1 binding to DLL4-Intron3 and very little recruitment of EP300 or ERG, which suggests that FOXO1 occupancy at this enhancer prevents EP300 and ERG binding or recruitment.
The DLL4-Intron3 enhancer does not contain a known FOXO1 binding site (Fig 4I). One hypothesis is that FOXO1 binds directly to RBPJk at this enhancer site [64]. To test this, we made a DLL4-Intron3 luciferase construct containing a mutated RBPJk site. When stimulated with VEGF, the DLL4-Intron3-RBPJkmut construct does not have a significant difference in luciferase activity compared to the wild type DLL4-Intron3 (Fig 4K). However, when FOXO1 is overexpressed, paradoxically, the luciferase activity of the wild type DLL4-Intron3 is significantly increased (Fig 4K). This suggests that FOXO1 not only regulates transient DLL4 transcriptional activity, as inhibition of FOXO1 allows for the burst in DLL4 expression in response to VEGF, but is also required to maintain baseline DLL4 expression previously observed when FOXO1 is knocked down (Fig S4C). Maintenance of DLL4 expression is perhaps achieved by FOXO1 binding to RBPJk, as mutation of the RBPJk site in the DLL4-Intron3 enhancer significantly reduced FOXO1-dependent induction of enhancer activity (Fig 4K).
FOXO1 inhibition restores microvascular regeneration in vivo
Phosphorylation of FOXO1 by AKT signaling allows for the export of FOXO1 from the nucleus, thereby inactivating its transcription factor activity [31]. To test whether this mechanism holds true in vivo, we analyzed kidney tissue from mice treated with bpV for the presence of total and Phospho-FOXO1 in endothelial cells during regeneration using automated morphometry (Fig 5A-B). BpV treatment enhanced p-FOXO1 in the endothelium 5 days following the nephrectomy-IRI surgery (Fig 5B), providing a clear in vivo link between enhanced AKT and enhancing p-FOXO1. Moreover, in that model of injury/repair in the kidney, bpV treatment is trending towards decreasing FOXO1 target genes Cited2 and Spry1 (Fig 5C) and enhancing Dll4 (Fig S2C). However, we only observed a decrease in Spry1 protein levels (Fig 5D and S5A), but not Cited2 protein levels (Fig S5B-C). In the following experiments, we tested whether FOXO1 inhibition in vivo could improve microvascular regeneration after ischemia reperfusion injury of the kidney. The FOXO1 inhibitor was given to mice 2 days before and 2 days after enhanced p-FOXO1 was noted in the bpV treated animals after injury, during the regenerative phase of the disease (Fig 5E). Inhibitor was administered orally until Day 7, at doses sufficient to achieve 1.5 × IC50 coverage (Fig S5D). At Day 7, although levels of FOXO1 transcripts were unaffected, FOXO1 target genes, Cited2 and Spry1 were suppressed by approximately 50% (Fig 6F). Consistent with these observations, we detected reduced protein levels of Spry1 (Fig 5H and S5E) but not Cited2 (Fig S5F-G). Next, we evaluated the effect of FOXO1 inhibition on transcripts for Apln, Kdr, Dll4, all of which have been implicated in tip cell formation (Fig 5G). These genes were upregulated by FOXO1 inhibition (Fig 5G). In addition, Plvap, a microvascular endothelium specific gene was enhanced consistent with increased regeneration of the vasculature (Fig 5G). More importantly, vascular coverage of LTL+ proximal tubule cells was significantly increased (Fig 5I-J) in animals treated with the FOXO1 inhibitor. These benefits, translated into an improvement in kidney function at Day 7, such as a reduction in urine albumin levels (Fig 5K) and a decrease in BUN (Fig S5H), suggesting that direct regeneration of the microvasculature can improve organ function.
Fig 5. FOXO1 inhibition in vivo enhances angiogenesis, revascularization and organ function during injury/repair.
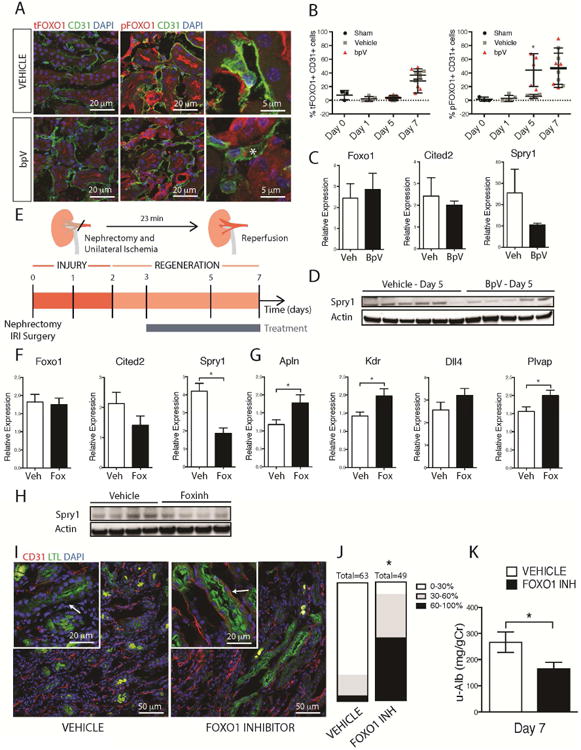
(A-B) Fluorescence images and (B) quantification of total and P-FOXO1 in mouse kidneys after IRI in the presence of bpV or vehicle. Note increased P-FOXO1 in endothelium in the presence of bpV day 5 after IRI (C). Q-PCR of kidneys d5 after IRI showing the effect of bpV on transcript levels (D) Whole tissue western blots showing levels of Sprouty1 in the presence of bpV (E) Schema and timecourse showing injury/repair caused by IRI in mouse kidney, the phase of regeneration and the duration of treatment with FOXO1 inhibitor (F) Q-PCR results showing the effect of FOXO1 inhibition on whole tissue levels of target genes Cited2 and Spry1 at day 5 after injury (G) Q-PCR showing the effect of FOXO1 inhibition on levels of angiogenesis and endothelial markers in kidney day 7 after IRI including the VEGF-A dependent FOXO1 regulated genes, such as Apln, Kdr, and Dll4. (H) Western blot showing whole tissue levels of Sprouty1 d5 after injury (I-J) Fluorescence images and quantification of endothelial density in the kidney following IRI. Note the preservation of capillaries around proximal tubules in kidneys treated with FOXO1 inhibitor. Graph shows proportion of kidney with tubules that have differing levels of capillary coverage (K) Urinary Albumin/Creatinine ratio at 7d after IRI from mice treated with FOXO1 inhibitor compared to vehicle. (n = 5 animals/group, *P < 0.05)
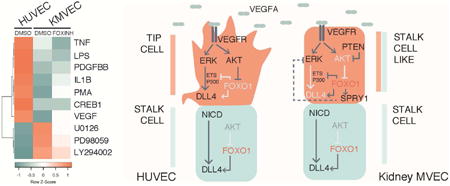
Fig 6. Model incorporating the role of FOXO1 in sprouting angiogenesis.
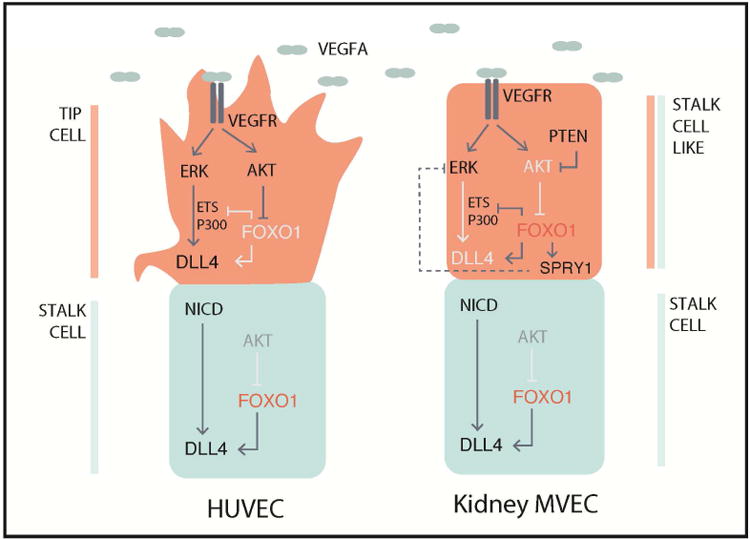
Induction of sprouting angiogenesis traditionally studied in HUVECs (left side) occurs through the activation of the VEGFR by ligand binding, which elicits a signaling cascade to activate DLL4 expression through two major signaling pathways, MAPK/ERK and PI3K/AKT. ERK signaling serves to increase ETS and EP300 recruitment to activate expression of DLL4, and AKT serves to inhibit FOXO1 activity allowing for the recruitment of transcriptional activators. Expression of DLL4 in turn activates Notch signaling in neighboring stalk cells, which has reduced VEGF-induced AKT signaling, resulting in high FOXO1 activity. High FOXO1 activity in the stalk cells serves to maintain DLL4 expression and prevents ETS/EP300 recruitment to Intron 3 of DLL4. Kidney MVECs (right side) however are desensitized to VEGF induced signaling due to hyperactive PTEN thereby inhibiting phosphorylation and activation of AKT, resulting in high FOXO1 activity even in the presence of VEGF. FOXO1 inhibits recruitment of transcriptional activators ETS and EP300 to Intron 3 of DLL4. In addition, FOXO1 may regulate the expression of the ERK inhibitor SPRY1 which may further impact ETS and EP300 activities. Altogether, this results in a lack of tip cell fate and reduced angiogenic capacity.
Discussion
In many tissues the angiogenic response to destruction of capillaries occurring during tissue injury results in restoration of the capillary bed. Similar studies in mouse and human kidney disease, however, indicate this is not the case [50, 65, 66]. Although a mild angiogenic response occurs within a few days following acute kidney injury, all models of chronic kidney disease are ultimately characterized by vascular regression without significant vascular regeneration [38, 67]. Accumulating evidence indicates loss of the kidney microvasculature may be a central or early problem in many forms of kidney disease [55]. Capillary loss leading to ischemia of the nephron, in critical areas of high metabolic demand, are now well documented experimentally [66]. Ischemic tubules of the nephrons respond by releasing inflammatory factors, recruiting leukocytes, exhibiting abnormal salt and water handling, impaired secretion of uremic toxins, setting up the kidney for chronic inflammation, fibrosis and hypertension [55, 68]. Considering that the success of tissue engineering relies heavily on the presence of a functional vasculature, understanding the mechanisms why capillaries fail to regenerate, could be a major advance toward preventing the development of chronic diseases directly, or toward designing alternative strategies and biomaterials to enhance tissue regeneration.
Although VEGFR2 is the cardinal receptor controlling angiogenesis, signaling from this receptor has pleiotropic effects on endothelial cell function. Multiple intracellular signaling cascades are activated upon receptor-ligand ligation, but how these pathways respectively control angiogenesis has been unclear. We have built upon the current knowledge to show the AKT and ERK pathways co-operate to control angiogenesis, demonstrating that the duration of the ERK signal, the translocation of p-ERK to the nucleus are critical aspects of ERK signaling that enable angiogenesis. Moreover, disruption of either pathway has a profound effect on successful angiogenesis. Directly downstream of ERK signaling is control of DLL4, a ligand for Notch signaling and instrumental in tip cell formation [15, 69, 70]. Successful signaling via PI3K/AKT inactivates FOXO1 by phosphorylating it [31]. We show here that FOXO1 is bound to a highly conserved enhancer in Intron 3 of DLL4. We further show that ERK dependent transcriptional regulators implicated in DLL4 activation including ERG and EP300 [54] are no longer able to bind to this enhancer when FOXO1 is bound, thereby explaining its suppressive activity. Thus PI3K/AKT, via control of FOXO1 activity, can directly regulate ERK dependent DLL4 activation, providing a direct link between MAPK/ERK and PI3K/AKT signaling pathway during angiogenesis. Furthermore, we show that not only is FOXO1 involved in the inhibition of DLL4 expression in response to VEGF, but that it is also involved in maintaining DLL4 expression by binding to a transcriptional cofactor RBPJk. The Notch Intracellular Domain (NICD) has previously been shown to bind to RBPJk to maintain DLL4 expression during arterio-venous specification [54]. It is therefore possible that FOXO1 binds to this complex during angiogenesis to desensitize DLL4 to stimulation by VEGF-A signaling, such as in stalk cells. In tip cells, when VEGF-A signaling is active, this in turn activates AKT signaling to inhibit FOXO1 activity, allowing for ETS factors and EP300 transcription factors to transiently activate expression of DLL4. In KMVECs however, due to the hyperactive PTEN activity, AKT signaling is impaired, which allows FOXO1 to occupy the 3rd Intron in DLL4, resulting in the inhibition of ETS and EP300 recruitment and a stalk cell like phenotype with little sprouting angiogenesis (Fig 6). These results illustrate that inhibition of FOXO1 in KMVECs allows the cells to be more responsive to VEGF and thereby allowing for classical tip-stalk selection at the vascular front. However, whether FOXO1 affects the speed at which tip cell selection occurs, or whether it regulates other aspects of tip cell selection is unclear and thus warrants further research.
Next, we show that in endothelial cells, FOXO1 has other critical target genes that suppress ERK signaling, most notably SPRY1. SPRY1, which is upregulated by FOXO1 in the presence of VEGF-A, has been shown to suppress ERK signaling directly by downregulating p-ERK [61]. Here we showed that in the presence of FOXO1 inhibitor, p-ERK localizes more readily to the nucleus. Whether this is due to SPRY1 inactivity remains to be explored. To this end, FOXO1 has broader suppressive activity on ERK signaling in the endothelium. We identified additional targets that are suppressed by FOXO1: these include CYR61 and SOCS3, both of which have been implicated in regulating angiogenesis and VEGF signaling [71, 72].
The insights into control of VEGFR2 signaling demonstrated here have derived from the study of endothelium that exhibits differential capacity to undergo angiogenesis. Why might these tissue-specific endothelial beds behave so differently? Firstly, recent fate-mapping studies indicating MVECs derive from local tissue-specified mesenchyme, and in the CNS from pial and periventricular membranes, rather than from the yolk sac blood islands, from where large vessel endothelium originates [73-75]. Therefore cell-autonomous tissue-specific endothelial cell programing during development may exist and aid to regulate vascular patterning. Secondly, the large vessels are coated with smooth muscle cells that suppress angiogenesis, but upon wounding can revascularize organs (Fig 1). Thirdly unlike skin, which is prone to wounding and tissue repair, solid organs are less prone to injury. Finally, internal organs have specific physiological environments including low oxygen tension, high osmolar concentrations, requirements for low permeability co-efficient, which require suppression of angiogenesis. These tissue-specific environments often have high VEGF levels that may permit MVEC survival. The consequence of physiologically driven suppression of angiogenesis appears to be impaired capacity to regenerate microvascular networks by angiogenesis after tissue injury. This suppression is increasingly recognized to contribute to morbidity and mortality especially in age-related diseases. Indeed, failure to regenerate the vasculature after acute or repetitive low grade injury may underlie many chronic diseases driven by environmental factors.
Conclusions
These studies identify FOXO1 as a critical link between VEGFR2/KDR signaling and angiogenesis. Here we showed that FOXO1 directly regulates DLL4 and Notch signaling in response to VEGFR2 activation. DLL4 and Notch are the cardinal-ligand receptor pair controlling tip and stalk cell formation during initiation of angiogenesis [15, 69, 70]. FOXO1, upstream regulators of FOXO1 activity, as well as downstream targets of FOXO1, can be targeted to benefit productive angiogenesis and vascular regeneration after tissue injury, and hence form the basis for novel therapeutics that can selectively regulate regeneration in vascular beds where this process is impeded.
Supplementary Material
Table 1. QPCR Primer List.
| Gene Name | TaqMan Probe | Species |
|---|---|---|
| KDR | Hs00911700_m1 | Human |
| EGR3 | Hs00231780_m1 | Human |
| EGR1 | Hs00152928_m1 | Human |
| CDKN2B | Hs00793225_m1 | Human |
| SERPINE | Hs00299953_m1 | Human |
| HEY1 | Hs01114113_m1 | Human |
| APLN | Hs00175572_m1 | Human |
| NDN | Hs00267349_s1 | Human |
| CITED2 | Hs01897804_s1 | Human |
| SPRY1 | Hs01083036_s1 | Human |
| ANGPT2 | Hs00169867_m1 | Human |
| HMOX1 | Hs01110250_m1 | Human |
| SPRY4 | Hs01935412_s1 | Human |
| DLL4 | Hs00184092_m1 | Human |
| FLT1 | Hs01052961_m1 | Human |
| NRP1 | Hs00826128_m1 | Human |
| NRP2 | Hs00187290_m1 | Human |
| GAPDH | Hs02786624_g1 | Human |
| Gapdh | Mm99999915_g1 | Mouse |
| Apln | Mm00443562_m1 | Mouse |
| Kdr | Mm01222421_m1 | Mouse |
| Dll4 | Mm00444619_m1 | Mouse |
Acknowledgments
We wish to thank Moody Mohamed, Sara Simpson and Thomas Neumann from Nortis Inc. for assistance. In addition, we would like to thank the following people at Biogen for assistance and guidance: Allie Roach, Matt Flegal, Norm Allaire, Chris Roberts, and Birgit Obermeier. These studies were funded by Biogen, Cambridge, MA; the National Institutes of Health [grant numbers DK093493, DK094768]; and the American Heart Association [grant number 12040023]. T.A. received funding from Daiiko-Sanchyo to study as a visiting Scientist at the University of Washington.
Footnotes
Lead contact: Lan T. H. Dang
Conflicts of Interest: Employees of Biogen have company stock. J.S.D has patents for therapies to stimulate organ regeneration.
Author contributions: LD, TA, GL, GM, IG, JSD developed the concepts and designed experiments. LD, TA, GL, AH, GM, BJ, SR, CS performed in vitro experiments and generated primary cells. LD, GM, AH, SA, CY, CC and JSD designed and performed microfluidics experiments. AA and RN performed organ explant experiments. LD, TA, JSD, BJ, NN, SR performed in vivo experiments. LD designed and made reporter assays. LD designed and performed ChIP assays. LD and SS performed bioinformatics analyses. LD and JSD wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ye L, et al. Transplantation of nanoparticle transfected skeletal myoblasts overexpressing vascular endothelial growth factor-165 for cardiac repair. Circulation. 2007;116(11 Suppl):I113–20. doi: 10.1161/CIRCULATIONAHA.106.680124. [DOI] [PubMed] [Google Scholar]
- 2.Kolesky DB, et al. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv Mater. 2014;26(19):3124–30. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 3.Kolesky DB, et al. Three-dimensional bioprinting of thick vascularized tissues. Proc Natl Acad Sci U S A. 2016;113(12):3179–84. doi: 10.1073/pnas.1521342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra R, et al. Effect of prevascularization on in vivo vascularization of poly(propylene fumarate)/fibrin scaffolds. Biomaterials. 2016;77:255–66. doi: 10.1016/j.biomaterials.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brudno Y, et al. Enhancing microvascular formation and vessel maturation through temporal control over multiple pro-angiogenic and pro-maturation factors. Biomaterials. 2013;34(36):9201–9. doi: 10.1016/j.biomaterials.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim DK, et al. Selective binding of C-6 OH sulfated hyaluronic acid to the angiogenic isoform of VEGF(165) Biomaterials. 2016;77:130–8. doi: 10.1016/j.biomaterials.2015.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moulisová V, et al. Engineered microenvironments for synergistic VEGF – Integrin signalling during vascularization. Biomaterials. 2017;126:61–74. doi: 10.1016/j.biomaterials.2017.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraehenbuehl TP, et al. Cell-responsive hydrogel for encapsulation of vascular cells. Biomaterials. 2009;30(26):4318–24. doi: 10.1016/j.biomaterials.2009.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baranski JD, et al. Geometric control of vascular networks to enhance engineered tissue integration and function. Proc Natl Acad Sci U S A. 2013;110(19):7586–91. doi: 10.1073/pnas.1217796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khademhosseini A, Langer R. A decade of progress in tissue engineering. Nat Protoc. 2016;11(10):1775–81. doi: 10.1038/nprot.2016.123. [DOI] [PubMed] [Google Scholar]
- 11.Silva EA, Mooney DJ. Effects of VEGF temporal and spatial presentation on angiogenesis. Biomaterials. 2010;31(6):1235–41. doi: 10.1016/j.biomaterials.2009.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herbert SP, Stainier DY. Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat Rev Mol Cell Biol. 2011;12(9):551–64. doi: 10.1038/nrm3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerhardt H. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin M, et al. Vegfa signals through ERK to promote angiogenesis, but not artery differentiation. Development. 2016;143(20):3796–3805. doi: 10.1242/dev.137919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445(7129):781–4. doi: 10.1038/nature05577. [DOI] [PubMed] [Google Scholar]
- 16.Hellstrom M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445(7129):776–80. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 17.Seib FP, et al. Multifunctional silk-heparin biomaterials for vascular tissue engineering applications. Biomaterials. 2014;35(1):83–91. doi: 10.1016/j.biomaterials.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basile DP, et al. Renal ischemia reperfusion inhibits VEGF expression and induces ADAMTS-1, a novel VEGF inhibitor. Am J Physiol Renal Physiol. 2008;294(4):F928–36. doi: 10.1152/ajprenal.00596.2007. [DOI] [PubMed] [Google Scholar]
- 19.Chiba T, Hukriede N, de Caestecker MP. Kidney Regeneration: Lessons from Development. Curr Pathobiol Rep. 2015;3(1):67–79. doi: 10.1007/s40139-015-0069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamoto Y, Tokunaga H, Tomita K. Vascular endothelial growth factor is an essential molecule for mouse kidney development: glomerulogenesis and nephrogenesis. Journal of Clinical Investigation. 1997;99(10):2351–2357. doi: 10.1172/JCI119416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, et al. The Accumulation of VEGFA in the Glomerular Basement Membrane and Its Relationship with Podocyte Injury and Proteinuria in Alport Syndrome. PLoS One. 2015;10(8):e0135648. doi: 10.1371/journal.pone.0135648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eremina V, et al. Glomerular-specific alterations of VEGF-A expression lead to distinct congenital and acquired renal diseases. J Clin Invest. 2003;111(5):707–16. doi: 10.1172/JCI17423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin SL, et al. Targeting endothelium-pericyte cross talk by inhibiting VEGF receptor signaling attenuates kidney microvascular rarefaction and fibrosis. Am J Pathol. 2011;178(2):911–23. doi: 10.1016/j.ajpath.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang BH, et al. Phosphatidylinositol 3-kinase signaling mediates angiogenesis and expression of vascular endothelial growth factor in endothelial cells. Proc Natl Acad Sci U S A. 2000;97(4):1749–53. doi: 10.1073/pnas.040560897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang B, et al. A dynamic H3K27ac signature identifies VEGFA-stimulated endothelial enhancers and requires EP300 activity. Genome Res. 2013;23(6):917–27. doi: 10.1101/gr.149674.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Val S, et al. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135(6):1053–64. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randi AM, et al. Regulation of angiogenesis by ETS transcription factors. Biochem Soc Trans. 2009;37(Pt 6):1248–53. doi: 10.1042/BST0371248. [DOI] [PubMed] [Google Scholar]
- 28.Craig MP, et al. Etv2 and fli1b function together as key regulators of vasculogenesis and angiogenesis. Arterioscler Thromb Vasc Biol. 2015;35(4):865–76. doi: 10.1161/ATVBAHA.114.304768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, et al. Both FOXO3a and FOXO1 are involved in the HGF-protective pathway against apoptosis in endothelial cells. Cell Biol Int. 2015 doi: 10.1002/cbin.10486. [DOI] [PubMed] [Google Scholar]
- 30.Wilhelm K, et al. FOXO1 couples metabolic activity and growth state in the vascular endothelium. Nature. 2016;529(7585):216–20. doi: 10.1038/nature16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potente M, et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. 2005;115(9):2382–92. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furuyama T, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279(33):34741–9. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- 33.Milkiewicz M, et al. Identification of a mechanism underlying regulation of the anti-angiogenic forkhead transcription factor FoxO1 in cultured endothelial cells and ischemic muscle. Am J Pathol. 2011;178(2):935–44. doi: 10.1016/j.ajpath.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang F, et al. Genetic engineering of human stem cells for enhanced angiogenesis using biodegradable polymeric nanoparticles. Proc Natl Acad Sci U S A. 2010;107(8):3317–22. doi: 10.1073/pnas.0905432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen RR, et al. Spatio-temporal VEGF and PDGF delivery patterns blood vessel formation and maturation. Pharm Res. 2007;24(2):258–64. doi: 10.1007/s11095-006-9173-4. [DOI] [PubMed] [Google Scholar]
- 36.Kida Y, et al. EphrinB2 reverse signaling protects against capillary rarefaction and fibrosis after kidney injury. J Am Soc Nephrol. 2013;24(4):559–72. doi: 10.1681/ASN.2012080871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ligresti G, et al. A Novel Three-Dimensional Human Peritubular Microvascular System. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2015070747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemos DR, et al. Maintenance of vascular integrity by pericytes is essential for normal kidney function. Am J Physiol Renal Physiol. 2016;311(6):F1230–f1242. doi: 10.1152/ajprenal.00030.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagashima T, et al. Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol. 2010;78(5):961–70. doi: 10.1124/mol.110.065714. [DOI] [PubMed] [Google Scholar]
- 40.Schmid AC, et al. Bisperoxovanadium compounds are potent PTEN inhibitors. FEBS Lett. 2004;566(1-3):35–8. doi: 10.1016/j.febslet.2004.03.102. [DOI] [PubMed] [Google Scholar]
- 41.Ligresti G, et al. The acute phase reactant orosomucoid-1 is a bimodal regulator of angiogenesis with time- and context-dependent inhibitory and stimulatory properties. PLoS One. 2012;7(8):e41387. doi: 10.1371/journal.pone.0041387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ligresti G, et al. Macrophage-derived tumor necrosis factor-alpha is an early component of the molecular cascade leading to angiogenesis in response to aortic injury. Arterioscler Thromb Vasc Biol. 2011;31(5):1151–9. doi: 10.1161/ATVBAHA.111.223917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bayless KJ, Davis GE. Sphingosine-1-phosphate markedly induces matrix metalloproteinase and integrin-dependent human endothelial cell invasion and lumen formation in three-dimensional collagen and fibrin matrices. Biochem Biophys Res Commun. 2003;312(4):903–13. doi: 10.1016/j.bbrc.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 44.Ren S, et al. LRP-6 is a coreceptor for multiple fibrogenic signaling pathways in pericytes and myofibroblasts that are inhibited by DKK-1. Proc Natl Acad Sci U S A. 2013;110(4):1440–5. doi: 10.1073/pnas.1211179110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staropoli JF, et al. Rescue of gene-expression changes in an induced mouse model of spinal muscular atrophy by an antisense oligonucleotide that promotes inclusion of SMN2 exon 7. Genomics. 2015;105(4):220–8. doi: 10.1016/j.ygeno.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Allaire NE, et al. Optimization of a high-throughput whole blood expression profiling methodology and its application to assess the pharmacodynamics of interferon (IFN) beta-1a or polyethylene glycol-conjugated IFN beta-1a in healthy clinical trial subjects. BMC Res Notes. 2013;6:8. doi: 10.1186/1756-0500-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 48.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duffield JS, et al. Restoration of tubular epithelial cells during repair of the postischemic kidney occurs independently of bone marrow-derived stem cells. J Clin Invest. 2005;115(7):1743–55. doi: 10.1172/JCI22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrimpf C, et al. Pericyte TIMP3 and ADAMTS1 modulate vascular stability after kidney injury. J Am Soc Nephrol. 2012;23(5):868–83. doi: 10.1681/ASN.2011080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomez IG, et al. Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J Clin Invest. 2015;125(1):141–56. doi: 10.1172/JCI75852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tourovskaia A, et al. Tissue-engineered microenvironment systems for modeling human vasculature. Exp Biol Med (Maywood) 2014;239(9):1264–71. doi: 10.1177/1535370214539228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chau BN, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med. 2012;4(121):121ra18. doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wythe JD, et al. ETS factors regulate Vegf-dependent arterial specification. Dev Cell. 2013;26(1):45–58. doi: 10.1016/j.devcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fligny C, Duffield JS. Activation of pericytes: recent insights into kidney fibrosis and microvascular rarefaction. Curr Opin Rheumatol. 2013;25(1):78–86. doi: 10.1097/BOR.0b013e32835b656b. [DOI] [PubMed] [Google Scholar]
- 56.Li B, et al. Mobilized human hematopoietic stem/progenitor cells promote kidney repair after ischemia/reperfusion injury. Circulation. 2010;121(20):2211–20. doi: 10.1161/CIRCULATIONAHA.109.928796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin P, et al. Wound healing in the PU.1 null mouse--tissue repair is not dependent on inflammatory cells. Curr Biol. 2003;13(13):1122–8. doi: 10.1016/s0960-9822(03)00396-8. [DOI] [PubMed] [Google Scholar]
- 58.Mirza R, DiPietro LA, Koh TJ. Selective and specific macrophage ablation is detrimental to wound healing in mice. Am J Pathol. 2009;175(6):2454–62. doi: 10.2353/ajpath.2009.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273(22):13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 60.Brunet A, et al. Akt Promotes Cell Survival by Phosphorylating and Inhibiting a Forkhead Transcription Factor. Cell. 1999;96(6):857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 61.Hanafusa H, et al. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4(11):850–8. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- 62.Impagnatiello MA, et al. Mammalian sprouty-1 and -2 are membrane-anchored phosphoprotein inhibitors of growth factor signaling in endothelial cells. J Cell Biol. 2001;152(5):1087–98. doi: 10.1083/jcb.152.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen DH, et al. Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc Natl Acad Sci U S A. 2013;110(17):6712–7. doi: 10.1073/pnas.1221526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kitamura T, et al. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J Clin Invest. 2007;117(9):2477–85. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lerman LO, Chade AR. Angiogenesis in the kidney: a new therapeutic target? Curr Opin Nephrol Hypertens. 2009;18(2):160–5. doi: 10.1097/MNH.0b013e32831ec1db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schrimpf C, et al. The role of pericyte detachment in vascular rarefaction. J Vasc Res. 2014;51(4):247–58. doi: 10.1159/000365149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ligresti G, et al. A Novel Three-Dimensional Human Peritubular Microvascular System. J Am Soc Nephrol. 2016;27(8):2370–81. doi: 10.1681/ASN.2015070747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campanholle G, et al. Cellular mechanisms of tissue fibrosis. 3. Novel mechanisms of kidney fibrosis. Am J Physiol Cell Physiol. 2013;304(7):C591–603. doi: 10.1152/ajpcell.00414.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hellstrom M. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 70.Leslie JD. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134:839–844. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- 71.Sun Y, et al. SOCS3 in retinal neurons and glial cells suppresses VEGF signaling to prevent pathological neovascular growth. Sci Signal. 2015;8(395):ra94. doi: 10.1126/scisignal.aaa8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Y, et al. CCN1/Cyr61-PI3K/AKT signaling promotes retinal neovascularization in oxygen-induced retinopathy. Int J Mol Med. 2015;36(6):1507–18. doi: 10.3892/ijmm.2015.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vasudevan A, et al. Compartment-specific transcription factors orchestrate angiogenesis gradients in the embryonic brain. Nat Neurosci. 2008;11(4):429–39. doi: 10.1038/nn2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng T, et al. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature. 2013;500(7464):589–92. doi: 10.1038/nature12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mugford JW, et al. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008;324(1):88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


