Figure 2.
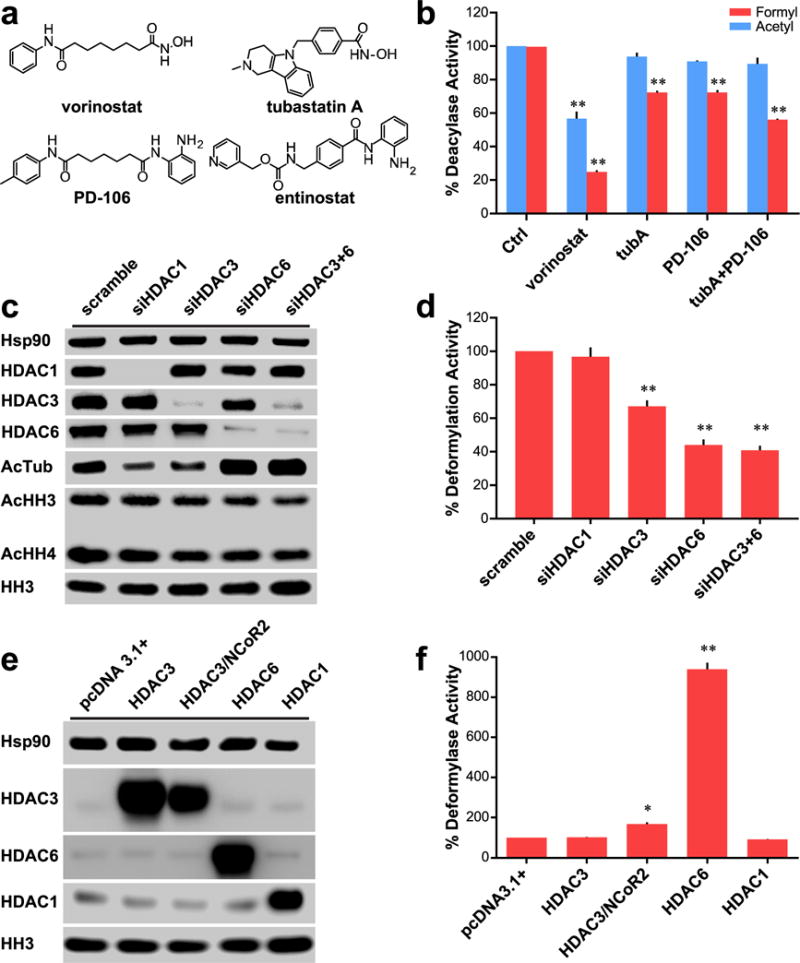
Role of HDACs 3 and 6 in cellular deformylation. (a) Chemical structures of small molecule inhibitors of HDACs 1, 2, 3, and 6. (b) Comparison of effect on deformylase and deacetylase activities of Hek293 lysates with various inhibitors for 24 h; data normalized to control deacylase activity. Vorinostat and PD-106 used at 1 μM; tubA used at 0.5 μM. (c) Western blot analysis of Hek293 cells with various siRNA transfections demonstrating selective and specific knockdown of targeted HDACs. (d) Deformylase activity of siRNA treated Hek293 lysates; data normalized to scramble deacetylase activity. (e) Western blot analysis of Hek293 cells transfected with various vectors to overexpress targeted HDACs. (f) Deformylase activity of transfected Hek293 cells; data normalized to pcDNA3.1+ vector transfected cells. Parts b, d, and f are n = 3; error bars are SEM. Parts c and e are representative of n ≥ 2 experiments. *p-value < 0.01. **p-value < 0.0001. All statistical analyses were Dunnett’s multiple comparisons of means to the means of their respective controls.
