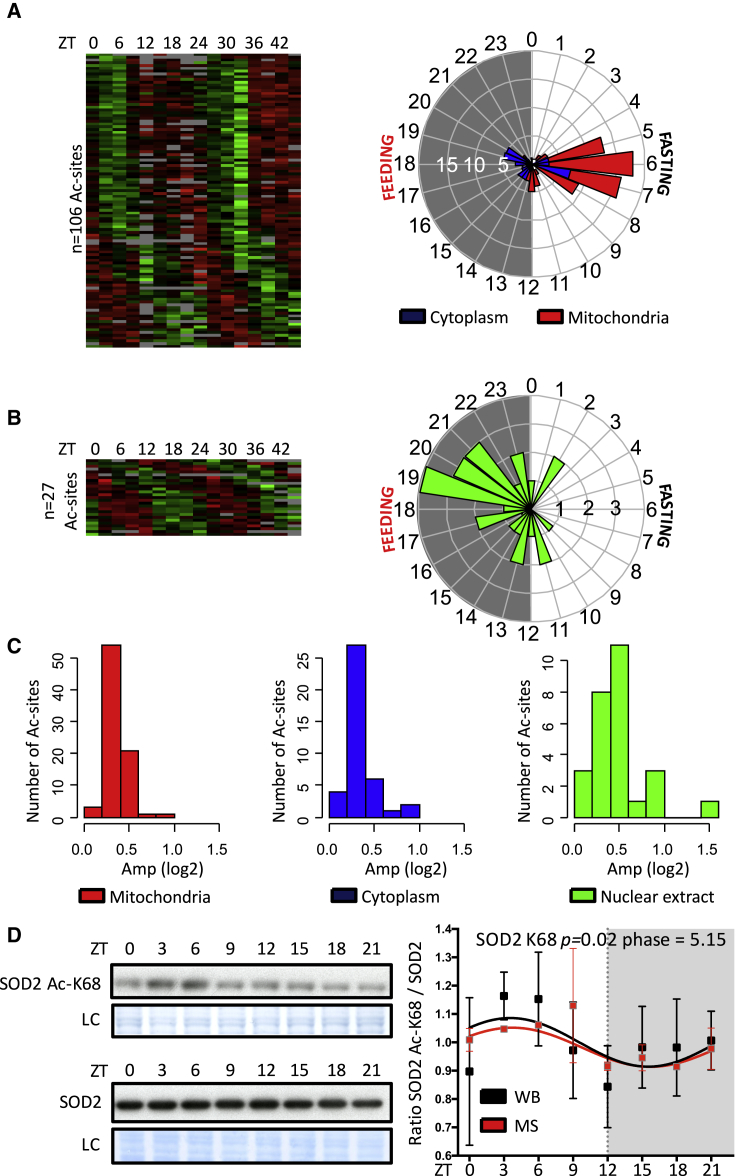
Figure 2.
Rhythmicity Analysis of the Liver Acetylome
(A–C) Heatmaps showing rhythmic acetylation sites normalized by their corresponding total protein amount in TE (A) and NE (B) under light/dark and night-restricted feeding conditions. Data were standardized by rows, and gray blocks indicate missing protein data. The polar plots on the right of each heatmap display peak phase distribution of the rhythmic acetylation (Ac) sites, and (C) shows amplitude. The colors of the polar plots (A and B) and histograms (C) indicate Ac sites that have a corresponding total protein with a defined mitochondrial (n = 80 Ac sites, red) or cytoplasmic (n = 40 Ac sites, blue) localization or Ac sites that are identified in NE (n = 27 Ac sites, green).
(D) Western blot (WB) analysis of total protein extracts using acetyl-K68 SOD2 (top blot) or total SOD2 (bottom blot). MS data in red, mean ± SEM, are from Table S2. WB data in black, mean ± SEM, are from three independent biological samples and represent the ratio of the acetyl-K68 SOD2 signal on the total SOD2 signal. Data (MS and WB) are normalized to the temporal mean. Naphtol blue black staining of the membranes was used as a loading control (LC) and served as a reference for normalization of the quantified values.