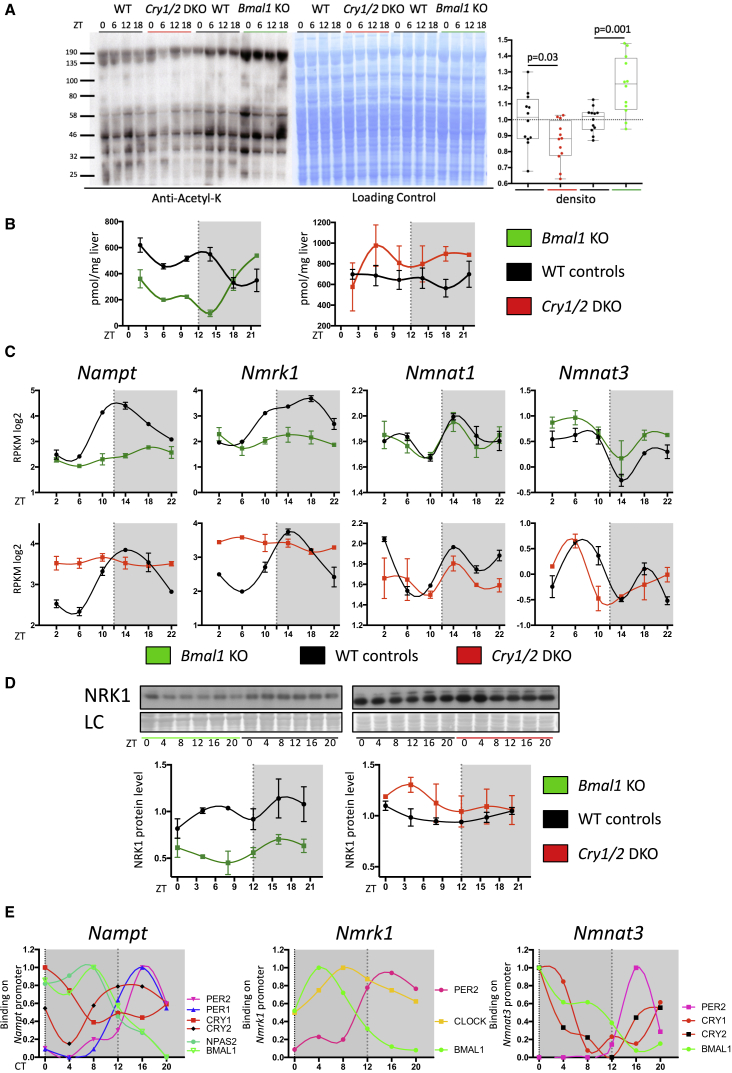
Figure 4.
Regulation of Global Acetylation and NAD+ Metabolism by the Circadian Clock
(A) Western blot analysis of global lysine acetylation performed on total protein extract from Cry1/2 DKO, Bmal1 KO, and their corresponding WT control mice (left). Naphtol blue black staining of the membrane was used as a loading control (center) and served as a reference for normalization of the quantified values displayed at the right. The average acetylation level was compared between clock-disrupted mice and their controls using Student’s t test (n = 12/genotype). Error bars represent min to max.
(B) NAD+ liver concentrations in Bmal1 KO (n = 2/time point) and Cry1/2 DKO mice (n = 2/time point) and their corresponding WT controls (mean ± SEM).
(C) Top: mRNA expression levels of Nampt, Nmrk1, Nmnat1, and Nmnat3 in Bmal1 KO mice (green) and their WT littermates (black) under night-restricted feeding conditions. Bottom: the same results in Cry1/2 DKO (red) mice and their WT controls (black). The data are from Atger et al. (2015); n = 2 for each time point and each genotype). Error bars represent ±SEM.
(D) WB analysis in TE extracts of NRK1 in Bmal1 KO and Cry1/2 DKO mice. Quantifications of the blots are displayed below. Naphtol blue black staining of the membranes was used as a loading control and served as a reference for normalization of the quantified values. Error bars represent ±SEM.
(E) Temporal binding of circadian clock core regulators on the Nampt, Nmrk1, and Nmnat3 gene promoters. At each time point, for each factor, the averages of all rhythmic bindings are presented. The data are from Koike et al. (2012).