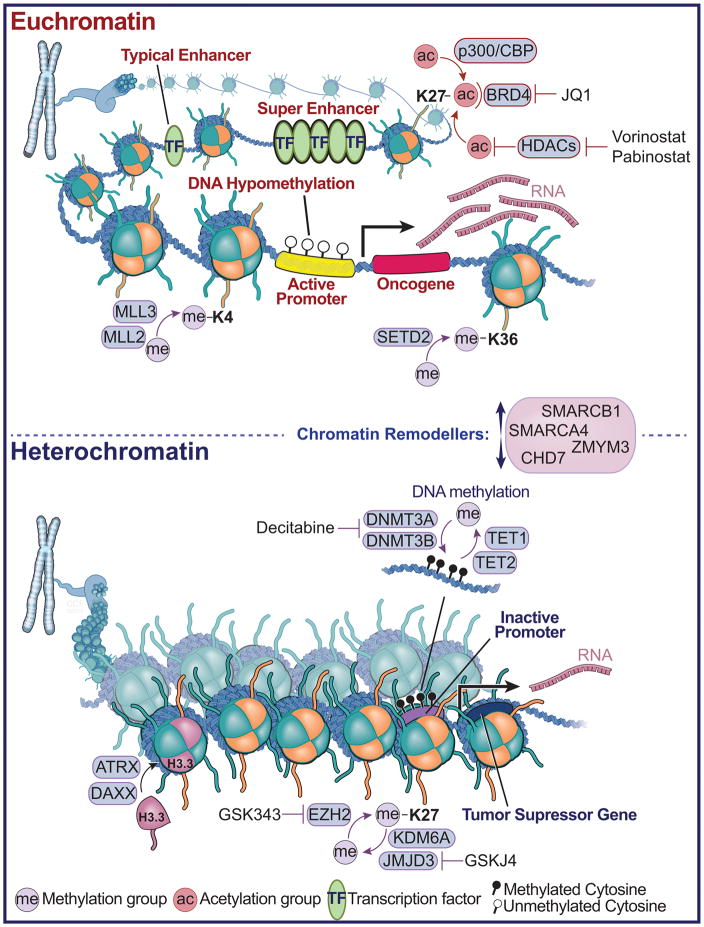
Figure 2. Brain Tumors Converge on Chromatin Architecture.
A diagram depicting euchromatin and histone modifications that mediate ‘active’ transcription in cancer cells (top panel). Shown are various histone modifications and enzymes, which catalyze the addition of post-translational modifications such as histone methylation and acetylation, or bind to these modifications, such as the BRD4, which binds acetylated lysine residues on histones. The green ovals represent transcription factor binding sites and locations of enhancers, or clusters of enhancers, termed super-enhancers. Also shown are drug compounds, which inhibit the removal (Vorinostat and Pabinostat - Histone deacetylase (HDAC) inhibitor) or detection of acetylation (JQ1). Shown in the middle are the chromatin remodelers, which facilitate the landscape of higher order chromatin structure towards euchromatin or heterochromatin. In the lower panel, heterochromatin is depicted and the associated modifications that mediate tumor suppressor gene silencing. These include the DNA methyltransferase family of enzymes, which catalyze the addition of methyl groups to cytosine - guanine di-nuclueotides, and TET enzymes, which facilitate DNA de-methylation through 5-methyl cytosine hydroxylation. Also shown is EZH2, which methylates at histone H3 at the 27th position, and the associated histone H3K27 de-methylases KDM6A and JMJD3. Shown are chemical inhibitors that reverse the methylation marks deposited or removed by these methyltransferase and demethylase enzymes related to heterochromatin (Decitabine, GSK343, GSKJ4). ATRX and DAXX are depicted which function to incorporate the histone H3.3 variant, and which are frequently mutated in pediatric high-grade glioma.