Figure 2.
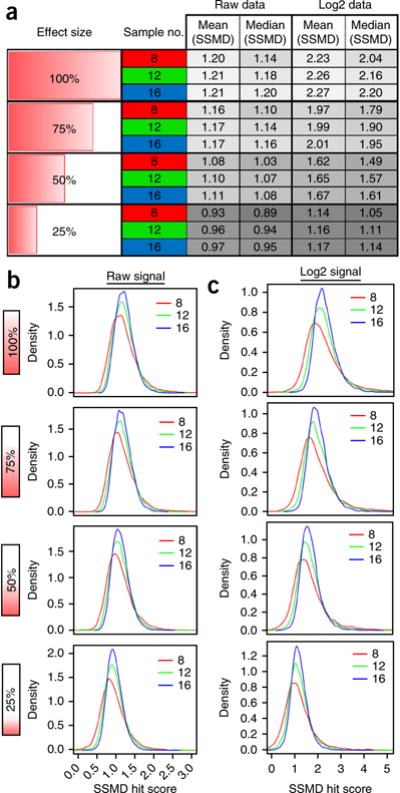
Predicted SSMD scores using bootstrapping. (a and b) Computational random sampling with replacement (‘bootstrapping’) of positive- and negative-control data sets provides a means of estimating HTS assay performance via a series of ‘virtual’ experiments at a defined sample size. This strategy is useful for establishing predicted SSMD scores relative to effect size, particularly for in vivo HTS assays in which same-day follow-up is desirable (e.g., visual phenotyping of ‘hit’ plates following ARQiv). Real-time data outputs (Fig. 5) facilitate same-day follow-up, but bootstrapping is necessary to establish reasonable approximations of hit criteria before initiating the full-scale screen. (a) Predicted SSMD scores (gray columns, right) were generated via 10,000 bootstrap iterations of raw and log-transformed control data. SSMD hit estimates were calculated relative to effect size (left column) and at three different sample numbers (8, 12, and 16; red, green, and blue, respectively, middle column). The results show SSMD estimates decreasing with smaller effect sizes and demonstrate the expanded dynamic range that log-transforming data provide. Note that for both raw and log-transformed data sets, lowering the sample number decreases the SSMD estimate (mean/median). (b) Density plots of bootstrap iterations using raw data show how increasing sample size reduces SSMD variance. Comparisons across effect size show a gradual decrease in predicted SSMD average as effect size is reduced. (c) Density plots of bootstrap iterations using log-transformed data also show reduced variance with larger sample sizes. Comparisons across effect size show the expanded SSMD score range following log transformation.
