Abstract
This study aimed to verify the reliability of the 7 tissue differentially methylated regions used in the methylated DNA immunoprecipitation (MeDIP) real- time quantitative polymerase chain reaction (real-time qPCR) based approach of fetal DNA in maternal blood to diagnosis of fetal trisomy 21. Forty pregnant women with high risk pregnancy who were referred after first or second trimester screening tests, were selected randomly. For each sample whole DNA extraction (mother and fetus), fragmentation of DNA, immunoprecipitation of methylated DNA and real- time qPCR using 7 primer pairs was performed. D-value for each sample was calculated using the following formula D = -4.908+ 0.254 XEP1+ 0.409 XEP4+ 0.793 XEP5+ 0.324 XEP6+ 0.505 XEP7+ 0.508 XEP9+ 0.691 XEP12. In all normal cases, D value was negative, while it was positive in all trisomy cases. Therefore, all normal and trisomy 21 cases were classified correctly which correspond to 100% specificity and 100% sensitivity for this method. The MeDIP real-time qPCR method has provided the opportunity for noninvasive prenatal diagnosis of fetal trisomy 21 to be potentially employed into the routine practice of diagnostic laboratories.
Key Words: Down syndrome, trisomy 21, prenatal diagnosis, cell-free fetal DNA; noninvasive prenatal testing; prenatal genetic screening
Down syndrome (DS) is the most prevalent genetic disease worldwide affecting about one in every 750 live births in all populations and is considered to be the most frequent etiology of intellectual disability (1-4). DS is caused by trisomy of whole or part of chromosome is accompanied with a large amount of health and social costs for patients and their families (5). It is coupled with many health issues, including mental retardation, congenital heart defects, gastrointestinal anomalies, audiovestibular and visual impairment, hematop-oietic disorders, early-onset Alzheimer disease and many other health problems (1, 2, 6, 7). While most fetal aneuplordy leads to miscarriage, trisomy 21 has the maximum survival rate. Due to the highest survival rate, the prenatal detection of fetal trisomy 21 is one of the commonest reasons for referral of women for prenatal diagnosis (8, 9). The incidence of births of children with DS rises with the age of the mother. Screening for DS is an important part of routine prenatal care. Prenatal diagnosis was presented in the 1970s with the major aim of detecting common aneuploidies such as trisomies 21, 18 and 13 (7, 10). Screening tests like the first trimester combination test are nowadays accessible to all pregnant women. Risk calculation is based on maternal age, nuchal translucency (NT) measurement by sonography and two serum markers: free beta hCG (free β-hCG) and pregnancy-associated plasma protein-A (PAPP-A). The test properties are rather good with a detection rate of almost 85– 90% with a false-positive rate of 5–9% (9, 11).
Following a positive prenatal screening test, women are usually recommended to perform fetal karyotyping, which is considered as the gold standard to confirm the presence or absence of aneuploidies. Despite that, the main problem of karyotyping is the long period of time needed to achieve definitive results. Other faster and cheaper methods which have been introduced include interphase FISH and QF-PCR, but their main disadvantage is that they do not provide a full graphic demonstration of all chromosomes (12-15). Prenatal genetic diagnosis of DS and other aneuploidies is done using common cytogenetic tests or DNA analysis which needs fetal DNA to be obtained by invasive methods such as chorionic villus sampling (CVS) during the first trimester and amniocentesis during the second trimester (16-18). Even so, all these methods are invasive and associated with risk of fetal loss (12, 19, 20). Therefore, developing a reliable technique for noninvasive prenatal diagnosis (NIPD) of fetal trisomy 21 is very important (8, 21, 22).
Over the last few years, a great quantity of investigation have been accomplished on the development of noninvasive prenatal testing (NIPT) for fetal aneuploidies (23, 24). The discovery of cell free fetal DNA (cffDNA) in the maternal circulation has opened up a new horizon in the field of prenatal care and screening. Detection of chromosomal aneuploidy is a challenging goal in NIPD research (22, 25-27). Due to the high maternal DNA background and the nature of cffDNA in maternal plasma, determination of chromosomes dosage in the fetal genome is very difficult by common methods. To overcome these issues, background maternal DNA interference can be diminished by using molecular signatures present in maternal plasma but originating completely from fetus. The discovery of fetal-specific DNA methylation signatures in maternal blood offered an excellent opportunity to advent and improve new approaches for noninvasive screening testing. Genes that show differential DNA methylation between placental tissues and maternal blood cells have been used as fetal nucleic acid markers (20, 28-31). DNA methylation is a dynamic process and could change during development. It is believed that more than half of tissue differentially methylated regions (TDMRs) are methylated in embryonic tissues and during the differentiation, they undergo de-methylation process.These TDMRs have been used to enrich and assess fetal DNA ratio by using monoclonal antibody for methylated CpGs using MeDIP approach (16, 18, 32, 33).
The aim of this study was to evaluate and validate the methylated DNA immunoprecipitation of fetal DNA in maternal blood for diagnosis of fetal trisomy 21.
Materials and methods
Sample collection and processing
The samples included 40 pregnant women referred between October 2014 to December 2015 to the Medical Genetic Center of Genome (Iran, Isfahan). All pregnant women agreed to participate in the study and signed an informed consent. The study was approved by the Bioethics Committee of Isfahan University of Medical Sciences. The experimental procedure was followed as previously described with some modifications (16, 33). The participants were women with singleton pregnan-cies, between 13 and 21 weeks of gestation. All participants underwent invasive prenatal diagnosis by CVS or amniocentesis followed by FISH or chromosomal analysis. Briefly, for each pregnant woman 4 ml of peripheral blood was collected on EDTA and then aliquoted into four 1.5 ml tubes and stored at −80°C within 4 h of collection until further use.
Extraction and fragmentation of DNA
DNA was extracted from 400 l of peripheral blood sample via QIAamp DNA blood mini kit (QIAGEN) according to the manufacturer's instruction. Subsequently the DNA was quantified using a UV spectrophotometer at 260 and 280 nm and 5 g of the DNA was sheared by sonication at 100% power for 20 min using a WiseClean WUC Digital Ultrasonic (WUC-D06H) into fragment sizes of 100 to 500 bp. Verification of sheared DNA was done by electrophoresis on 1.5% agarose gel.
MeDIP-real time qPCR
Sonicated DNA was processed using the MeDIP methodology for immunoprecipitation of hypermethylated fragments (Diagenode’s MagMeDIP kit) according to the manufacturer’s instruction. Finally, real-time qPCR was carried out on an input and immunoprecipitated fragments for the selected 7 differentially methylated regions (DMRs) on chromosome 21 and 2 control regions (hypermethylated region on chromosome 13 and hypomethylated region on chromosome 22) as described previously (33). The real- time PCR was performed with specific primers (Table 1) and Maxima SYBR Green/ ROX qPCR Master Mix (Thermo Fisher Scientific) using StepOne Plus real-time PCR system (Applied Biosystems, USA). Amplification conditions were: first denaturation and enzyme activation at 95 C for 10 min, followed by 40 cycles of amplification at 95 C for 15 s and 60 C for 1 min. The reactions were performed in triplicate. Initially 6 maternal peripheral blood samples with known karyotype (normal pregnancies) were used to calculate the median for normalized □Ct. The ratio value for each of the DMRs was calculated using the median of normalized □Ct obtained from known samples. Finally, D value amount for each unknown sample was calculated using the following formula (33):
Table 1.
Primers used for real-time PCR (16)
| Region | Primer Name | Sequence | Amplicon size (bp) |
|---|---|---|---|
| EP1 | EP1-F | 5’-GCTGGACCAGAAAGTGTTGAG-3’ | 149 |
| EP1-R | 5’-GTGTGCTGCTTTGCAATGTG-3’ | ||
| EP4 | EP4-F | 5’-CTGTTGCATGAGAGCAGAGG-3’ | 95 |
| EP4-R | 5’-CGTCCCCCTCGCTACTATCT-3’ | ||
| EP5 | EP5-F | 5’-TGCAGGATATTTGGCAAGGT-3’ | 127 |
| EP5-R | 5’-CTGTGCCGGTAGAAATGGTT-3’ | ||
| EP6 | EP6-F | 5’-TGAATCAGTTCACCGACAGC-3’ | 104 |
| EP6-R | 5’-GAAACAACCTGGCCATTCTC-3’ | ||
| EP7 | EP7-F | 5’-CCGTTATATGGATGCCTTGG-3’ | 127 |
| EP7-R | 5’-AAACTGTTGGGCTGAACTGC-3’ | ||
| EP9 | EP9-F | 5’-GACCCAGACGATACCTGGAA-3’ | 110 |
| EP9-R | 5’-GCTGAACAAAACTCGGCTTC-3’ | ||
| EP12 | EP12-F | 5’-ATTCTCCACAGGGCAATGAG-3’ | 128 |
| EP12-R | 5’-TTATGTGGCCTTTCCTCCTG-3’ | ||
| HYP113c | HYP113 c-F | 5’-CAGGAAAGTGAAGGGAGCTG-3’ | 79 |
| HYP113 c-R | 5’-CAAAACCCAATGGTCAATCC-3’ | ||
| U122d | U122 d-F | 5’-AAGGTGCCCAATTCAAGGTA-3’ | 104 |
| U122 d-R | 5’-CTTCCCCACCAGTCTTGAAA-3’ |
ΔCTPB Normal= CTPBNormal Input- CTPBNormal IP
ΔCTPB T21=CTPBT21Input- CTPB T21IP
Where IP correspond to Immunoprecipitated and PB represent the peripheral blood.
Norm ΔCT value PBNormal= EΔCTPB Normal
Norm ΔCT value PB T21= EΔCTPB T21
Where E= 10[-1/slope] and Norm= Normalized
Ratio value sample; DMR= Norm ΔCTPB Sample(Normal or T21)/Median (Norm ΔCTPB Normal)
D= -4.908+ 0.254 XEP1+ 0.409 XEP4+ 0.793 XEP5+ 0.324 XEP6+ 0.505 XEP7+ 0.508 XEP9+ 0.691 XEP12
where XEPi is fraction value for each EP marker (33).
Results
The ratio values obtained from the 7 selected DMRs (Table 2) were applied to the prediction equation for each sample separately to calculate the D value and determine their status (normal or trisomy 21) (Tables 3 to 5). Cases that gave a D value above zero (cutting point) were classified as “trisomy 21” and below zero was classified as “normal”. A total of 26 cases were classified as normal whereas the remaining 14 cases were classified as trisomy 21 (Table 3). The ratio values and D values obtained for normal and trisomy 21 samples were compared in Table 4. Statistical evaluation of the diagnostic efficiency of the discriminant analysis function, using this method showed a perfect classification for all normal and trisomy 21 cases resulting in a sensitivity and specificity of 100%. Karyotyping of the samples confirmed the above findings (Table 2).
Table 2.
Ratio values obtained for 40 samples
| Sample | Status | EP1 | EP4 | EP5 | EP6 | EP7 | EP9 | EP12 |
|---|---|---|---|---|---|---|---|---|
| 1 | TRISOMY | 1.398713031 | 1.926523788 | 1.33403822 | 6.712266515 | 0.054477337 | 0.749292024 | 1.12537046 |
| 2 | TRISOMY | 80.81213255 | 8.975547844 | 19.84770098 | 26.97591518 | 0.701687224 | 0.037296912 | 4.588110062 |
| 3 | NORMAL | 3.140730166 | 0.377879982 | 1 | 0.020964613 | 2.487850856 | 1.845101267 | 1 |
| 4 | NORMAL | 0.748775074 | 2.35169821 | 0.068878378 | 0.407612116 | 0.440901253 | 0.08029923 | 0.873795923 |
| 5 | TRISOMY | 3.812401332 | 1.994462503 | 1.468354179 | 1 | 1.329422798 | 1 | 1.12654114 |
| 6 | NORMAL | 0.000217214 | 0.000752408 | 0.000812978 | 0.00026385 | 0.000352914 | 0.000547636 | 0.000574424 |
| 7 | TRISOMY | 0.930836701 | 4.597979392 | 0.992060493 | 11.53703023 | 0.311412768 | 1.003610869 | 0.164709994 |
| 8 | TRISOMY | 2.186464614 | 1.486582984 | 1.929865094 | 21.499035 | 0.543706507 | 0.395349362 | 0.366021424 |
| 9 | TRISOMY | 3.143997347 | 1.094293701 | 1.055919608 | 13.54041452 | 2.960621374 | 0.799516659 | 4.254530692 |
| 10 | TRISOMY | 3.725928527 | 0.47467106 | 0.561944673 | 8.563496658 | 3.523233276 | 0.33662168 | 1.5888688 |
| 11 | TRISOMY | 8.745858457 | 0.371645746 | 1.929865094 | 21.499035 | 0.135926627 | 0.395349362 | 0.366021424 |
| 12 | TRISOMY | 2.156512383 | 1.829817797 | 2.526254524 | 5.095063052 | 2.545942789 | 0.730420924 | 2.047693801 |
| 13 | TRISOMY | 8.773980448 | 1.145676711 | 1.480208083 | 28.44099435 | 10.39198969 | 3.655325801 | 1 |
| 14 | NORMAL | 1.148300315 | 1 | 1.069102368 | 2.484232152 | 0.595387154 | 0.619810886 | 0.546957257 |
| 15 | NORMAL | 0.000108882 | 0.000159179 | 9.11287E-05 | 1.59383E-05 | 0.000591521 | 0.037645925 | 0.03926866 |
| 16 | NORMAL | 1 | 2.291194604 | 1.834898169 | 0.858208443 | 2.001386775 | 0.81966142 | 0.150235744 |
| 17 | NORMAL | 0.329739814 | 1.918661226 | 0.224082207 | 1.151249077 | 0.00070436 | 0.008151041 | 0.162318574 |
| 18 | NORMAL | 2.651116373 | 0.012242144 | 1.462564247 | 2.305692308 | 0.966539099 | 0.961394197 | 1.153485605 |
| 19 | TRISOMY | 1.318228073 | 3.487032958 | 1.631840441 | 28.58526976 | 3.74223459 | 6.765516134 | 3.732131966 |
| 20 | NORMAL | 2.740805592 | 2.07915887 | 1.158694309 | 1.247638519 | 1.094217853 | 1 | 0.668963777 |
| 21 | TRISOMY | 7.318763871 | 6.480027789 | 1.054456807 | 17.16264495 | 1.841651394 | 0.78089474 | 1.519924856 |
| 22 | NORMAL | 4.122730053 | 2.279946545 | 1.718917138 | 0.007584133 | 1.034475875 | 1 | 0.627201102 |
| 23 | NORMAL | 2.04854559 | 0.496890547 | 0.201730342 | 2.553011436 | 0.559767643 | 0.697952132 | 1.435944511 |
| 24 | NORMAL | 3.120116721 | 1.216722359 | 1.24444287 | 2.73435394 | 0.766788178 | 0.596750593 | 0.33844657 |
| 25 | TRISOMY | 2.103220341 | 0.927873476 | 0.511746062 | 13.63459555 | 1.946523821 | 0.395349362 | 0.33844657 |
| 26 | NORMAL | 0.758173533 | 1.598811661 | 0.76286517 | 0.748565259 | 2.709262157 | 0.027760936 | 0.398044049 |
| 27 | NORMAL | 0.466710555 | 1.112650121 | 0.195264285 | 4.767391153 | 0.011437377 | 0.015126269 | 0.489031737 |
| 28 | NORMAL | 0.570619122 | 0.812252396 | 1.858965505 | 1.971371945 | 0.990274229 | 0.525659321 | 0.947370071 |
| 29 | NORMAL | 0.003987 | 0.399426 | 2.562408 | 0.014733 | 0.249636 | 0.689776 | 1.793776 |
| 30 | NORMAL | 0.673896996 | 1.02313747 | 0.460253309 | 1.063780134 | 1.77509963 | 1.128338548 | 0.690158677 |
| 31 | NORMAL | 0.54699517 | 0.498615626 | 0.341628443 | 3.021363992 | 2.208244625 | 1.72931418 | 0.914465089 |
| 32 | NORMAL | 0.010803181 | 0.173378871 | 0.128647917 | 0.600484953 | 0.525550025 | 0.010958544 | 0.968618189 |
| 33 | NORMAL | 0.375425877 | 0.734584317 | 0.101145217 | 0.462395736 | 0.698290863 | 0.008345418 | 0.004100456 |
| 34 | NORMAL | 0.45081268 | 0.321078952 | 0.268036244 | 0.003491832 | 0.406098049 | 0.007229943 | 0.38315499 |
| 35 | NORMAL | 1.306402992 | 2.177994031 | 0.464097554 | 0.665356752 | 2.946291143 | 0.198223512 | 0.542990928 |
| 36 | NORMAL | 0.503687209 | 0.117358968 | 2.111839216 | 5.653718293 | 0.246883102 | 0.01382286 | 0.005727019 |
| 37 | NORMAL | 2.587215093 | 0.018161158 | 0.426465222 | 0.002518534 | 0.021345859 | 0.64743174 | 0.005143621 |
| 38 | NORMAL | 1.734355926 | 0.054598306 | 1.243580586 | 0.0000128382 | 0.007105244 | 1.690910149 | 0.0000690021 |
| 39 | NORMAL | 2.636090682 | 0.063592481 | 0.531263683 | 0.012097058 | 0.00041262 | 0.058834247 | 0.039609766 |
| 40 | NORMAL | 0.022411764 | 0.054296387 | 0.05390265 | 2.53818988 | 0.000628456 | 0.38151191 | 0.026571068 |
| Mean | 4.003145281 | 1.449434464 | 1.445470785 | 5.988551466 | 1.319358826 | 0.796127543 | 0.91060985 | |
| Median | 1.358470552 | 1.011568735 | 1.027228404 | 2.138532127 | 0.699989044 | 0.60828074 | 0.58707918 | |
Table 3.
The specification of samples
| Sample | Fetal Status |
Prediction Value
(D-Value) |
Gestational Weeks | Fetal Gender | Confirmed by |
|---|---|---|---|---|---|
| 1 | Trisomy | 0.65367 | 17 | Female | FISH |
| 2 | Trisomy | 47.31239 | 18 | Male | FISH |
| 3 | Normal | -0.27123 | 16 | Female | Amniocentesis |
| 4 | Normal | -2.70204 | 17 | Female | Amniocentesis |
| 5 | Trisomy | 0.322288 | 17 | Male | Amniocentesis |
| 6 | Normal | -4.90605 | 16 | Female | Amniocentesis |
| 7 | Trisomy | 2.51462 | 18 | Male | Amniocentesis |
| 8 | Trisomy | 5.479775 | 16 | Female | Amniocentesis |
| 9 | Trisomy | 6.403729 | 17 | Male | FISH |
| 10 | Trisomy | 2.500866 | 20 | Female | Amniocentesis |
| 11 | Trisomy | 6.483923 | 16 | Female | Amniocentesis |
| 12 | Trisomy | 3.113981 | 18 | Male | Amniocentesis |
| 13 | Trisomy | 55.8825 | 15 | Female | Amniocentesis |
| 14 | Normal | -1.56116 | 14 | Male | FISH |
| 15 | Normal | -4.86127 | 18 | Female | Amniocentesis |
| 16 | Normal | -0.45287 | 16 | Male | Amniocentesis |
| 17 | Normal | -3.37215 | 21 | Female | Amniocentesis |
| 18 | Normal | -0.5492 | 19 | Female | Amniocentesis |
| 19 | Trisomy | 15.31432 | 16 | Male | Amniocentesis |
| 20 | Normal | -0.51555 | 16 | Female | Amniocentesis |
| 21 | Trisomy | 8.37518 | 15 | Male | Amniocentesis |
| 22 | Normal | -0.09896 | 16 | Female | Amniocentesis |
| 23 | Normal | -1.56781 | 16 | Male | Amniocentesis |
| 24 | Normal | -0.82083 | 16 | Female | Amniocentesis |
| 25 | Trisomy | 2.24684 | 15 | Female | Amniocentesis |
| 26 | Normal | -1.55669 | 16 | Male | Amniocentesis |
| 27 | Normal | -2.28352 | 14 | Female | Amniocentesis |
| 28 | Normal | -0.89621 | 18 | Male | Amniocentesis |
| 29 | Normal | -0.99089 | 15 | Female | Amniocentesis |
| 30 | Normal | -1.6622 | 15 | Male | Amniocentesis |
| 31 | Normal | -0.68975 | 18 | Female | Amniocentesis |
| 32 | Normal | -3.59748 | 16 | Female | Amniocentesis |
| 33 | Normal | -3.92246 | 18 | Male | Amniocentesis |
| 34 | Normal | -3.97498 | 15 | Female | Amniocentesis |
| 35 | Normal | -1.13799 | 21 | Male | Amniocentesis |
| 36 | Normal | -1.08992 | 15 | Female | Amniocentesis |
| 37 | Normal | -3.56119 | 16 | Male | Amniocentesis |
| 38 | Normal | -2.59636 | 18 | Female | Amniocentesis |
| 39 | Normal | -3.72975 | 13 | Female | CVS |
| 40 | Normal | -3.8025 | 17 | Male | Amniocentesis |
| Min | -4.90605 | 13 | |||
| Max | 55.88245615 | 21 | |||
| MEAN | 3.581176892 | 16.61904762 | |||
| MEDIAN | -1.57612 | 15.24471 | |||
Table 4.
The comparison of ratio and D values obtained for normal and trisomy 21 samples
|
Normal
|
Trisomy 21
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Min | Max | Average | Median | Min | Max | Average | Median | |
| EP1 | 0.000109 | 4.12273 | 1.248103 | 0.748775 | 0.000108882 | 4.122730053 | 1.248102725 | 0.748775074 |
| EP4 | 0.000159 | 2.351698 | 0.858713 | 0.498616 | 0.000159179 | 2.35169821 | 0.858712697 | 0.498615626 |
| EP5 | 0.0000911 | 2.562408 | 0.796095 | 0.464098 | 9.11287E-05 | 2.562408 | 0.796095449 | 0.464097554 |
| EP6 | 0.0000128 | 5.653718 | 1.30727 | 0.748565 | 1.28382E-05 | 5.653718293 | 1.307270292 | 0.748565259 |
| EP7 | 0.000353 | 2.946291 | 0.842427 | 0.559768 | 0.000352914 | 2.946291143 | 0.842426773 | 0.559767643 |
| EP9 | 0.000548 | 1.845101 | 0.548169 | 0.525659 | 0.000547636 | 1.845101267 | 0.548168811 | 0.525659321 |
| EP12 | 0.000069 | 1.793776 | 0.526149 | 0.489032 | 6.90021E-05 | 1.793776 | 0.526148993 | 0.489031737 |
| D Value | -4.90605 | -0.09896 | -2.14400069 | -2.370295 | 0.322288 | 55.88245615 | 14.18725183 | 5.186462 |
Discussion
The development of a NIPD technique for fetal trisomy 21 without carrying risk for the pregnancy is a promising research area in prenatal diagnosis (21, 34). The major challenge for the development of NIPD using cffDNA is the limited amount and fragmented structure of cffDNA in the maternal circulation. Over past few years, significant advances have been made for the enrichment and analysis of cffDNA. Nonetheless, most of these techniques are time consuming, laborious or difficult to implement on a large scale (25, 34). Currently, two methods have been developed and validated with almost 100% accuracy. The first method is based on next generation sequencing and the other one is based on MeDIP real-time qPCR. Several reports have shown that application of MeDIP in combination with real-time qPCR using maternal peripheral blood permits prenatal noninvasive detection of trisomy 21. Papageorgiou et al. in 2011 showed that the methylation ratio of normal and trisomy 21 cases for 12 selected DMRs could diagnose 14 trisomy 21 and 26 normal cases indicate 100% specificity and 100% sensitivity of the approach (16). Tsalikiet et al. in 2012 confirmed and evaluated this technique for noninvasive prenatal diagnosis of trisomy 21 in larger scale on 175 samples with 99.2% specificity and 100% sensitivity of the approach (33). In another study performed on 10 samples in 2013, Gorduza et al. showed that this approach could detect trisomy 21 cases with 100% specificity (19). The results of our research are in line with those of previous studies and corroborate the high sensitivity and specificity of this method for detection of trisomy 21.The main advantage of this method compared to the next generation sequencing technology is that, this approach could be accessible in all basic diagnostic laboratories as it requires no major and exclusive infrastructure, and is technically easier and less expensive. Moreover, this method will be able to offer results in less than 3 working days (16).
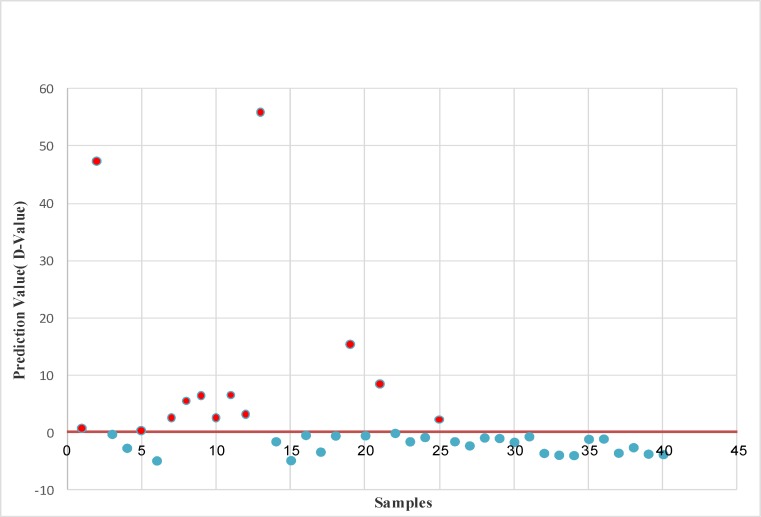
Fig. 1.
Prediction values (D) derived from the application of the diagnostic formula v1.1 for 40 blind tested cases. The Y axis represents the prediction value D and the X axis shows samples
As different ethnic groups may have different DNA methylation patterns and this could influence MeDIP-qPCR results, the main purpose of the present research was to evaluate and assess an optimized condition for NIPD of fetal trisomy 21 using cffDNA in maternal blood in Iran. We found that our results were in accordance with previous studies and this method is usable for screening in Iran. Furthermore, many different studies will need to be implemented to support the introduction of a new diagnostic strategy into the clinical practice of prenatal diagnostic laboratories.
This approach has provided the opportunity for NIPD of fetal trisomy 21 into many diagnostic laboratories (19, 35). Although the present study is based on a small sample of participants and data from more samples will be of help, our results confirm that this technology could be effective for screening trisomy 21 in pregnant women and could be applied in clinical practice.
Conflict of Interest
The authors declared no conflict of interest.
References
- 1.Antonarakis SE, Lyle R, Dermitzakis ET, et al. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet. 2004;5:725–38. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- 2.Leonard H, Wen X. The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev. 2002;8:117–34. doi: 10.1002/mrdd.10031. [DOI] [PubMed] [Google Scholar]
- 3.Lyle R, Bena F, Gagos S, et al. Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur J Hum Genet. 2009;17:454–66. doi: 10.1038/ejhg.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asim A, Kumar A, Muthuswamy S, et al. "Down syndrome: an insight of the disease". J Biomed Sci. 2015;22:41. doi: 10.1186/s12929-015-0138-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang X, Liu C, Yu T, et al. Genetic dissection of the Down syndrome critical region. Hum Mol Genet. 2015;24:6540–51. doi: 10.1093/hmg/ddv364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khocht A, Yaskell T, Janal M, et al. Subgingival microbiota in adult Down syndrome periodontitis. J Periodontal Res. 2012;47:500–7. doi: 10.1111/j.1600-0765.2011.01459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caban-Holt A, Head E, Schmitt F. Down Syndrome. Rosenberg’s Molecular and Genetic Basis of Neurological and Psychiatric Diseases. Fifth ed. 2015. pp. 163–70. [Google Scholar]
- 8.Lim JH, Park SY, Ryu HM. Non-invasive prenatal diagnosis of fetal trisomy 21 using cell-free fetal DNA in maternal blood. Obstet Gynecol Sci. 2013;56:58–66. doi: 10.5468/OGS.2013.56.2.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Go AT, van Vugt JM, Oudejans CB. Non-invasive aneuploidy detection using free fetal DNA and RNA in maternal plasma: recent progress and future possibilities. Hum Reprod Update. 2011;17:372–82. doi: 10.1093/humupd/dmq054. [DOI] [PubMed] [Google Scholar]
- 10.Loane M, Morris JK, Addor MC, et al. Twenty-year trends in the prevalence of Down syndrome and other trisomies in Europe: impact of maternal age and prenatal screening. Eur J Hum Genet. 2013;21:27–33. doi: 10.1038/ejhg.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang T, Dennis A, Meschino WS, et al. First trimester screening for Down syndrome using nuchal translucency, maternal serum pregnancy-associated plasma protein A, free-beta human chorionic gonadotrophin, placental growth factor, and alpha-fetoprotein. Prenat Diagn. 2015;35:709–16. doi: 10.1002/pd.4597. [DOI] [PubMed] [Google Scholar]
- 12.Gekas J, van den Berg DG, Durand A, et al. Rapid testing versus karyotyping in Down's syndrome screening: cost-effectiveness and detection of clinically significant chromosome abnormalities. Eur J Hum Genet. 2011;19:3–9. doi: 10.1038/ejhg.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar R, Singh P. Prenatal Diagnosis of Down Syndrome in a Molecular Era. Indian Journal of Genetics and Molecular Research. 2013;2:43–9. [Google Scholar]
- 14.Tekcan A, Tural S, Elbistan M, et al. The combined QF-PCR and cytogenetic approach in prenatal diagnosis. Mol Biol Rep. 2014;41:7431–6. doi: 10.1007/s11033-014-3630-7. [DOI] [PubMed] [Google Scholar]
- 15.Mann K, Ogilvie CM. QF-PCR: application, overview and review of the literature. Prenat Diagn. 2012;32:309–14. doi: 10.1002/pd.2945. [DOI] [PubMed] [Google Scholar]
- 16.Papageorgiou EA, Karagrigoriou A, Tsaliki E, et al. Fetal-specific DNA methylation ratio permits noninvasive prenatal diagnosis of trisomy 21. Nat Med. 2011;17:510–3. doi: 10.1038/nm.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolk H, Wellesley D. Antenatal screening for Down Syndrome and other chromosomal abnormalities: increasingly complex issues. Arch Dis Child Fetal Neonatal Ed. 2014;99:F2–3. doi: 10.1136/archdischild-2013-304384. [DOI] [PubMed] [Google Scholar]
- 18.Tsui DW, Lam YM, Lee WS, et al. Systematic identification of placental epigenetic signatures for the noninvasive prenatal detection of Edwards syndrome. PLoS One. 2010;5:e15069. doi: 10.1371/journal.pone.0015069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorduza EV, Popesca R, Caba L, et al. Prenatal diagnosis of 21 trisomy by quantification of methylated fetal DNA in maternal blood: study on 10 pregnancies. Rev Romana Med Lab. 2013;21:275–84. [Google Scholar]
- 20.Chim SSC. Potential application of fetal epigenetic markers on the non-invasive prenatal detection of chromosomal abnormality. Clin Chem Lab Med. 2014;52:585–8. doi: 10.1515/cclm-2014-0034. [DOI] [PubMed] [Google Scholar]
- 21.Old RW, Crea F, Puszyk W, et al. Candidate epigenetic biomarkers for non-invasive prenatal diagnosis of Down syndrome. Reproductive biomedicine online. 2007;15:227–35. doi: 10.1016/s1472-6483(10)60713-4. [DOI] [PubMed] [Google Scholar]
- 22.Chim SSC, Jin S, Lee TYH, et al. Systematic Search for Placental DNA-Methylation Markers on Chromosome 21: Toward a Maternal Plasma-Based Epigenetic Test for Fetal Trisomy 21. Clin Chem. 2008 Mar 1;54(3):500–11. doi: 10.1373/clinchem.2007.098731. [DOI] [PubMed] [Google Scholar]
- 23.van Schendel RV, Dondorp WJ, Timmermans DR, et al. NIPT-based screening for Down syndrome and beyond: what do pregnant women think? Prenat Diagn. 2015;35:598–604. doi: 10.1002/pd.4579. [DOI] [PubMed] [Google Scholar]
- 24.Drury S, Hill M, Chitty LS. Cell-Free Fetal DNA Testing for Prenatal Diagnosis. Adv Clin Chem. 2016;76:1–35. doi: 10.1016/bs.acc.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Latendresse G, Deneris A. An update on current prenatal testing options: first trimester and noninvasive prenatal testing. J Midwifery Womens Health. 2015;60:24–36. doi: 10.1111/jmwh.12228. quiz 111. [DOI] [PubMed] [Google Scholar]
- 26.Alix-Panabieres C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016;6:479–91. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 27.Alberry M, Maddocks D, Jones M, et al. Free fetal DNA in maternal plasma in anembryonic pregnancies: confirmation that the origin is the trophoblast. Prenat Diagn. 2007;27:415–8. doi: 10.1002/pd.1700. [DOI] [PubMed] [Google Scholar]
- 28.Chim SS, Tong YK, Chiu RW, et al. Detection of the placental epigenetic signature of the maspin gene in maternal plasma. Proc Natl Acad Sci U S A. 2005;102:14753–8. doi: 10.1073/pnas.0503335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–7. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 30.Hui WW, Chiu RW. Noninvasive prenatal testing beyond genomic analysis: what the future holds. Curr Opin Obstet Gynecol. 2016;28:105–10. doi: 10.1097/GCO.0000000000000252. [DOI] [PubMed] [Google Scholar]
- 31.Lee da E, Lim JH, Kim MH, et al. Novel Epigenetic Markers on Chromosome 21 for Noninvasive Prenatal Testing of Fetal Trisomy 21. J Mol Diagn. 2016;18:378–87. doi: 10.1016/j.jmoldx.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Papageorgiou EA, Fiegler H, Rakyan V, et al. Sites of differential DNA methylation between placenta and peripheral blood: molecular markers for noninvasive prenatal diagnosis of aneuploidies. Am J Pathol. 2009;174:1609–18. doi: 10.2353/ajpath.2009.081038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsaliki E, Papageorgiou EA, Spyrou C, et al. MeDIP real-time qPCR of maternal peripheral blood reliably identifies trisomy 21. Prenat Diagn. 2012;32:996–1001. doi: 10.1002/pd.3947. [DOI] [PubMed] [Google Scholar]
- 34.Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–75. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norwitz ER, Levy B. Noninvasive prenatal testing: the future is now. Rev Obstet Gynecol. 2013;6:48–62. [PMC free article] [PubMed] [Google Scholar]