Abstract
Background
Non-coding small RNAs (sRNAs) regulate a variety of important biological processes across all life domains, including bacteria. However, little is known about the functional evolution of sRNAs in bacteria, which might occur via changes in sRNA structure and/or stability or changes in interactions between sRNAs and their associated regulatory networks, including target mRNAs. The sRNA Pxr functions as a developmental gatekeeper in the model cooperative bacterium Myxococcus xanthus. Specifically, Pxr prevents the initiation of fruiting body development when nutrients are abundant. Previous work has shown that Pxr appears to have a recent origin within a sub-clade of the myxobacteria, which allowed us to infer the most recent common ancestor of pxr and examine the divergence of Pxr since its origin.
Results
To test for inter-specific divergence in functional effects, extant pxr homologs from several species and their inferred ancestor were introduced into an M. xanthus deletion mutant lacking pxr. Both the inferred ancestral pxr and all extant alleles from species containing only one copy of pxr were found to control development in M. xanthus in a qualitatively similar manner to the native M. xanthus allele. However, multiple paralogs present in Cystobacter species exhibited divergent effects, with two paralogs controlling M. xanthus development but two others failing to do so. These differences may have occurred through changes in gene expression caused by apparent structural differences in the sRNA variants encoded by these paralogs.
Conclusions
Taken together, our results suggest that Pxr plays a common fundamental role in developmental gene regulation across diverse species of myxobacteria but also that the functional effects of some Pxr variants may be evolving in some lineages.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-017-1037-5) contains supplementary material, which is available to authorized users.
Keywords: Bacterial development, Gene duplication, Multicellularity, Myxobacteria, Small non-coding RNAs, Regulation of gene expression
Background
Non-coding small RNAs are prevalent in regulating gene expression across all domains of life, from eukaryotes, archaea to bacteria [1–6] and include trans-encoded microRNAs (miRNAs) in animals and plants and small regulatory RNAs (sRNAs) in bacteria. These small RNAs modulate gene expression at the post-transcriptional level by base-pairing with their mRNA targets, resulting in mRNA degradation, inhibition of translation, or both. In bacteria, these small regulators play important roles in a variety of physiological processes, including stress responses [7–11], outer membrane protein synthesis [12–15], virulence [16] and social behaviors [7, 17, 18].
Bacterial sRNAs are often ~100 nucleotides in length (but can also be larger, e.g. [19, 20]), transcribed from the intergenic regions in bacterial genomes with their own promoters and commonly have multiple mRNA targets [2, 3]. They are predicted to fold into stable stem-loop structures including a ρ-independent terminator at the 3′ end. The function of an sRNA is determined by at least a seed region of ~6–8 bases that are complementary to a region of their mRNA targets, and this region is one of the most conserved features of homologous sRNA sequences [21, 22]. Even a single nucleotide change in this region can abolish sRNA-based regulation [7, 23–25], suggesting strong selective constraints on this region. Nevertheless, other sequence regions might also be prominent for function [23, 26].
Several proteins are known to participate in sRNA gene regulation. For example, many characterized sRNAs in enteric bacteria require the RNA chaperone Hfq for interactions with mRNA targets [27], although a number of sRNAs in other species require proteins other than Hfq [28, 29]. Ribonucleases also contribute to sRNA activities and levels. Most base-pairing sRNAs are transcribed as independent units, but few are processed in some manner [7, 30–32]. RNases E and RNase III also facilitate target mRNA degradation [33–35].
Recent studies have shown that the phylogenetic distributions and sequences of sRNAs tend to be lineage-specific [21, 36–39], suggesting independent origins of sRNAs in specific lineages followed by phylogenetically limited patterns of duplication, diversification and loss rather than a high rate lateral sRNA gene transfer across large phylogenetic distances [40]. For instance, a large fraction of Escherichia coli sRNAs has accumulated in the Enterobacteriales order since it split from the rest of the γ-proteobacteria [36].
Nevertheless, the functional evolution of sRNAs and how they coevolve with their associated regulatory networks remain poorly understood in most systems. In enteric bacteria, the SgrS sRNAs associated with glucose-phosphate stress appear to have coevolved with their associated networks, as many sgrS homologs from different species fail to control the same target mRNAs in E. coli that the respective E. coli-specific allele regulates [41]. However, sgrS is unique in that it also encodes a protein-coding gene sgrT, which regulates glucose-phosphate stress via an unknown mechanism [42]. It is thus unclear how representative sgrS may be of other sRNA systems with respect to evolutionary questions.
In this study, we sought to investigate evolution of an sRNA - Pxr - that controls fruiting body development in the myxobacteria. Many species of myxobacteria (δ-proteobacteria, order Myxococcales), including Myxococcus xanthus, survive starvation by multicellular development into fruiting bodies that yield stress-resistant spores [43]. In M. xanthus, Pxr functions as a developmental gatekeeper that prevents the initiation of fruiting body development when nutrients are abundant [7]. The pxr gene is located downstream of the σ54-dependent response regulator nla19, appears to be transcribed from an upstream σ54 promoter and is predicted to fold into a stable triple-stem-loop structure [7]. There are two pxr-specific sRNA forms, Pxr-L (long) and Pxr-S (short), and Pxr-S is thought to be the product of Pxr-L processing [7]. Both Pxr-L and Pxr-S are expressed at high levels during vegetative growth, but Pxr-S appears to be the primary negative regulator because it (but not Pxr-L) is rapidly and greatly diminished upon starvation, which presumably allows development to proceed [7]. Pxr appears to have a recent evolutionary origin in the lineage basal to the suborder Cystobacterineae within the Myxococcocales order [38]. Most species within this suborder contain a single copy of pxr; however, tandem paralogs were found within species of the genus Cystobacter [38].
Pxr was discovered from an evolution experiment in which a developmentally defective M. xanthus strain (‘OC’, obligate cheater) evolved into a strain with restored developmental proficiency (‘PX’, phoenix) due to a mutation that deactivated Pxr [44]. OC is a descendant of the developmentally proficient wild-type strain GJV1 and differs from GJV1 by 14 mutations that accumulated during laboratory evolution in a nutrient-rich liquid environment [45, 46]. This cheater strain is defective at development in clonal groups but can exploit GJV1 in chimeric groups to sporulate more efficiently than GJV1. Pxr-S, the primary regulatory form of Pxr, is constitutively expressed in OC, which in turn down-regulates development even when nutrients are depleted. Accordingly, OC is defective at fruiting body formation and sporulation on nutrient-free plates. Strain PX emerged from OC after a cheater-induced population crash [44]. PX differs from OC by a single C→A mutation in the first loop of Pxr, which abolishes the regulatory function of Pxr and confers the phenotype of fruiting body formation and high spore production even though the defective variant of Pxr-S continues to be produced [7, 44]. Taken together, these results suggested that Pxr is a negative regulator of M. xanthus development that blocks the initiation of fruiting body formation during vegetative growth. Indeed, deletion of pxr from the wild-type strain GJV1 allows development to proceed at high nutrient levels [7].
The fact that OC constitutively expresses Pxr-S provides an excellent opportunity to examine the functionality of pxr variants in its genetic background. Homologs of pxr proficient at blocking development in the OC background will demonstrate very low spore production, and vice versa. In this study, we first reconstructed the previously-inferred most recent common ancestor of extant pxr homologs and introduced it into an OC deletion mutant lacking pxr (hereafter “OC Δpxr”, which exhibits high sporulation like strain PX [44]) to test whether the pxr ancestral allele can restore the regulatory function of Pxr observed in M. xanthus. Next, we introduced pxr homologs from species with only one copy of this gene and other species with multiple pxr paralogs to test for functional divergence of the Pxr regulatory system in the myxobacteria.
Results
We first describe results with the inferred pxr ancestral allele and then results with extant homologs from different species to examine the functional divergence of Pxr since its origin in the myxobacteria. Nucleotide sequences of the inferred ancestor and extant homologs at every site are shown in Fig. 1a. As described in the Materials and Methods, for each constructed strain that carries a unique allele of pxr, we examined two or three independently isolated clones for their developmental phenotypes. Because no differences among independent clones of identical constructs were detected either within or among replicates, we used within-replicate means across independent clones as data points. For clarity, for the three control strains GJV1, OC and OC Δpxr, we retain their original designations. For the two isogenic control constructs that differ only in the presence or absence of the M. xanthus GJV1 pxr allele, we refer to them as “pxr GJV1” and “pxr null”. For constructs with different versions of pxr, we label them according to their strains of origin (e.g. “pxr Mxs33” for the pxr homolog from M. stipitatus strain Mxs33) or as “pxr Anc” for the inferred ancestral allele. Experimental strains of M. xanthus new to this study were derived from OC Δpxr and were generated by homologous integration of plasmids that carry the 429-nt region preceding pxr in M. xanthus in conjunction with different versions of pxr. A list of strains used is provided in Table 1 and the pxr mero-dioploid region of each constructed strain is shown on Fig. 2.
Fig. 1.
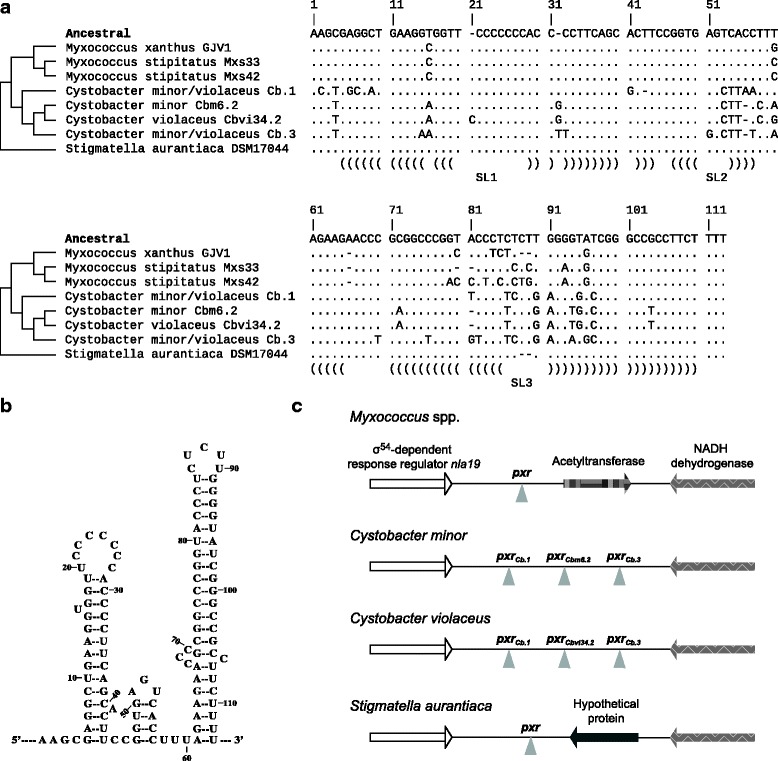
a Nucleotide sequences of the inferred pxr ancestor and extant homologs at every site from 5′ to 3′. A dot indicates a nucleotide identical to the corresponding site for the inferred ancestor at the internal node shared by the non-Stigmatella homologs, whereas letters show nucleotide differences. The cladogram of the pxr homologs is shown on the left. The consensus stem-loops based on the single-copy homologs are annotated as SL1, SL2 and SL3 at the bottom. Brackets represent nucleotides positioned on the stems. b The predicted secondary structure of the Pxr ancestor, which has a calculated self-folding free energy of −57.5 kcal/mol. The nucleotide numbers correspond to the alignment positions in (a). c Genetic organization of pxr alleles and their neighboring genes in different myxobacterial species. Gene and intergenic region lengths are not drawn to scale
Table 1.
List of M. xanthus strains and plasmids used in this study
| Strain | Genotype | pxr allele description | Reference/source |
|---|---|---|---|
| GJV1 | Derivative isolate of Myxococcus xanthus DK1622 | M. xanthus GJV1 pxr | [44, 47] |
| GVB207.3 | Evolutionary descendant of GJV1 (herein “OC”) | M. xanthus GJV1 pxr | [44, 46] |
| GJV207 | GVB207.3 Δpxr (herein “OC Δpxr”) | In-frame deletion of pxr | [7] |
| NY01 | GJV207 intergenic::pPxrGJV1, kanR | M. xanthus GJV1 pxr | This study |
| NY02 | GJV207 intergenic::pPxrnull, kanR | Intergenic region preceding pxr only | This study |
| KC00 | GJV207 intergenic::pPxrAnc, kanR | The inferred pxr ancestor shared by Myxococcus and Cystobacter spp. | This study |
| KC01 | GJV207 intergenic::pPxrMxs33, kanR | Myxococcus stipitatus Mxs 33 pxr | This study |
| KC02 | GJV207 intergenic::pPxrMxs42, kanR | Myxococcus stipitatus Mxs 42 pxr | This study |
| KC03 | GJV207 intergenic::pPxrCb.1, kanR | pxr Cb.1 in both Cystobacter minor Cbm 6 and Cystobacter violaceus Cbvi 34 | This study |
| KC04 | GJV207 intergenic::pPxrCbm6.2, kanR | pxr Cbm6.2 in C. minor Cbm 6 | This study |
| KC05 | GJV207 intergenic::pPxrCbvi34.2, kanR | pxr Cbvi34.2 in C. violaceus Cbvi 34 | This study |
| KC06 | GJV207 intergenic::pPxrCb.3, kanR | pxr Cb.3 in both C. minor Cbm 6 and C. violaceus Cbvi 34 | This study |
| KC07 | GJV207 intergenic::pPxrSga, kanR | Stigmatella aurantiaca DSM 17044 pxr | This study |
| Plasmid | Description | Reference/source | |
| pCR2.1 | Cloning vector | Invitrogen | |
| pPxrGJV1 | pCR2.1 with a 537-nt fragment that contains the 429-nt intergenic fragment and the 108-nt pxr coding region in M. xanthus | This study | |
| pPxrnull | pCR2.1 with only the 429-nt intergenic fragment in M. xanthus | This study | |
| pPxrAnc | pCR2.1 with the 429-nt intergenic fragment in conjunction with the inferred pxr ancestor shared by Myxococcus and Cystobacter. spp. | This study | |
| pPxrMxs33 | pCR2.1 with the 429-nt intergenic fragment in conjunction with the pxr allele from M. stipitatus strain Mxs 33 | This study | |
| pPxrMxs42 | pCR2.1 with the 429-nt intergenic fragment in conjunction with the pxr allele from M. stipitatus strain Mxs 42 | This study | |
| pPxrCb.1 | pCR2.1 with the 429-nt intergenic fragment in conjunction with pxr Cb.1 in both C. minor Cbm 6 and C. violaceus Cbvi 34 | This study | |
| pPxrCbm6.2 | pCR2.1 with the 429-nt intergenic fragment in conjunction with pxr Cbm6.2 in C. minor strain Cbm 6 | This study | |
| pPxrCbvi34.2 | pCR2.1 with the 429-nt intergenic fragment in conjunction with pxr Cbvi34.2 in C. violaceus strain Cbvi 34 | This study | |
| pPxrCb.3 | pCR2.1 with the 429-nt intergenic fragment in conjunction with pxr Cb.3 in both C. minor Cbm 6 and C. violaceus Cbvi 34 | This study | |
| pPxrSga | pCR2.1 with the 429-nt intergenic fragment in conjunction with the pxr allele from S. aurantiaca strain DSM 17044 | This study | |
“intergenic” refers to a 429-nt fragment that contains the 3′-terminal 167-nt of nla19 and the 262-nt intergenic region between nla19 and pxr in M. xanthus
Fig. 2.
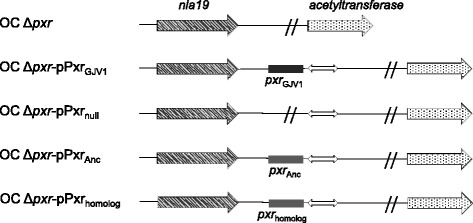
Genetic organization of the strain OC Δpxr and the constructed strains in this study. The double-arrowed blocks are integrated pCR2.1 vector fragments and the symbol // indicates the deletion of pxr
The inferred ancestral pxr allele negatively regulates development
The sequence of pxr ancestral to homologs within the Myxococcus and Cystobacter genera was previously inferred using the homolog from the distantly related genus Stigmatella as an outgroup (all three genera belong to the suborder Cystobacterineae in the order Myxococcocales) [38] (Fig. 1b). Although we refer to this allele as ancestral, it is not necessarily ancestral to the Stigmatella homolog and differs from it only by the presence of two bases (CU) in the third loop that are absent in the Stigmatella sequence. Between the inferred pxr ancestor and the extant M. xanthus GJV1 homolog there are seven base differences. Six of these differences are either near or on the third and last stem-loop of Pxr. Among them three are single base substitutions: a U→G change in the single-stranded region between the second and third stem-loops and a pair of complementary changes on the third stem (U→C combined with A→G). The other three differences are single base deletions: one at the base of the bulge on the left side of the third stem, one nearby the third loop and one in the loop. The seventh difference is a single base change U→C at the bulge on the left side of the first stem.
We examined the proficiency of the Pxr ancestor at controlling development in M. xanthus by measuring spore production on nutrient-free agar plates. PxrAnc blocked development just as effectively as PxrGJV1, as no spores at the lower limit of detection were obtained from the OC Δpxr-derived strain carrying either allele (Fig. 3a). This result indicates that the seven base differences between the ancestor and the M. xanthus copy do not affect the developmental phenotype assayed here. The Northern blot showed that pxr Anc was expressed into both Pxr-L and Pxr-S at similar levels and sizes as the extant M. xanthus allele (Fig. 3b).
Fig. 3.
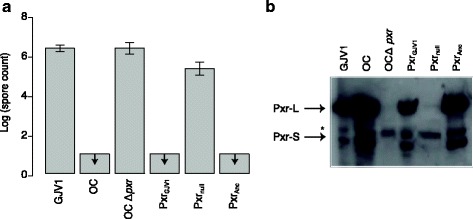
a The ancestral Pxr blocks M. xanthus development in a DK1622/GJV1-derived genomic background. Spore production of the OC Δpxr strain carrying the pxr ancestor and the control strains are shown. Arrows indicate that no spores were observed at the lower limit of detection. Error bars represent standard deviations. b Northern blot showing the expression of Pxr-L and Pxr-S produced by the OC Δ pxr strain carrying the pxr ancestor and the control strains. The asterisk marks binding of the probe to non-Pxr RNAs
pxr paralogs diverged greatly in sequence and functional effects
Seven previously identified pxr homologs were examined in this study. Three of them from phylogenetically diverse species (two from M. stipitatus strains Mxs33 and Mxs42 and one from Stigmatella aurantiaca DSM17044) are single-copy orthologs with a degree of sequence conservation similar to that of the 23S rRNA gene [38]. Four of them are paralogs found in Cystobacter minor Cbm 6 and C. violaceus Cbvi 34. Both of these strains carry multiple pxr paralogs in tandem (pxr Cb.1 in both strains, pxr Cbm6.2 in C. minor, pxr Cbvi34.2 in C. violaceus and pxr Cb.3 in both strains from 5′ to 3′) (Fig. 1c). The divergence among these paralogs within each Cystobacter species is greater than their orthologous divergence across species. For instance, the estimated evolutionary distances between pxr Cb.1 vs. pxr Cbm6.2, pxr Cbm6.2 vs. pxr Cb.3, and pxr Cb.1 vs. pxr Cb.3 in C. minor Cbm 6 are 14.4%, 17.4% and 22.6% (Table 2), but pxr Cb.1 and pxr Cb.3 in C. minor Cbm 6 and C. violaceus Cbvi 34 are identical to each other and pxr Cbm6.2 and pxr Cbvi34.2 only differ by two nucleotide positions across these two species. We thus infer that these paralogs originated by gene duplication in a common ancestor of C. minor and C. violaceus and diverged greatly from each other prior to the split between these species and that subsequent inter-specific divergence occurred only in the second paralogs.
Table 2.
Estimates of evolutionary distances between each pair of pxr homologs
| GJV1 | Mxs33 | Mxs42 | Cb.1 | Cbm6.2 | Cbvi34.2 | Cb.3 | Sga | |
|---|---|---|---|---|---|---|---|---|
| GJV1 | ||||||||
| Mxs33 | 0.064 | |||||||
| Mxs42 | 0.085 | 0.051 | ||||||
| Cb.1 | 0.322 | 0.285 | 0.421 | |||||
| Cbm6.2 | 0.233 | 0.246 | 0.351 | 0.144 | ||||
| Cbvi34.2 | 0.219 | 0.246 | 0.351 | 0.145 | 0.009 | |||
| Cb.3 | 0.344 | 0.282 | 0.417 | 0.226 | 0.174 | 0.189 | ||
| Sga | 0.076 | 0.052 | 0.113 | 0.219 | 0.179 | 0.179 | 0.298 |
The distances were determined using the maximum composite likelihood model implemented in MEGA v. 5.0 [67] and the numbers shown are base substitutions per site between sequences
All pxr homologs examined are located in the intergenic region between the σ54-dependent response regulator nla19 and a predicted NADH dehydrogenase gene (predicted genes Mxan_1078 and Mxan_1080 in the M. xanthus genome; [47]) but other neighboring gene content has diversified among genera (Fig. 1c). These homologs also appear to be transcribed from an upstream σ54 promoter like the one in M. xanthus and are predicted to fold into stable multi-stem-loop structures [38]. The two extant Myxococcus alleles, the Stigmatella allele and the second Cystobacter paralogs in both C. minor and C. violaceus (PxrCbm6.2 and PxrCbvi34.2) examined here are predicted to form three stem-loops, but the first and third Cystobacter paralogs (PxrCb.1 and PxrCb.3) are predicted to fold into only two long stem-loops and lack the second short stem-loop predicted for the other alleles [38]. The consensus secondary structure of Pxr based on the alignment of single-copy orthologs is indicated in Fig. 1a.
To ensure all pxr homologs are transcribed in the same manner as the native M. xanthus pxr allele, all the homologs in our constructs are preceded by the 429-nt region upstream of the pxr gene in M. xanthus (this region contains the 3′-terminal 167-nt of nla19 and the 262-nt intergenic region between nla19 and pxr). Despite extensive efforts to clone completely accurate sequences of pxr homologs, cloned sequences all lacked the first adenine. Because this adenine was absent from all cloned homologs we proceeded with these sequences for functional analysis. The developmental assays showed that the Myxococcus and Stigmatella orthologs (PxrMxs33, PxrMxs42 and PxrSga) all effectively blocked M. xanthus development, as the OC Δpxr-derived strains carrying them all produced almost no spores (Fig. 4a). These results demonstrate that nucleotides different between the orthologs tested are not essential for Pxr function and lacking the first adenine does not prevent effective blockage of development by Pxr.
Fig. 4.
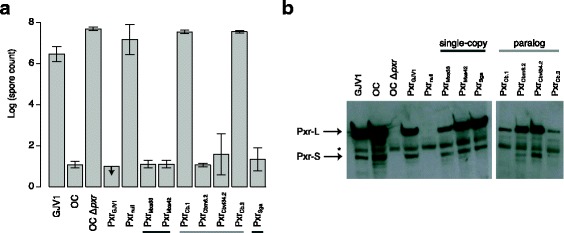
a Single-copy Pxr homologs from different species blocked M. xanthus development but there is large functional variation among the paralogs in Cystobacter. Spore production by OC Δpxr-derived strains carrying integrated pxr homologs from different myxobacterial species are shown. Dark grey bars indicate single-copy Pxr homologs and light grey bars indicate Pxr paralogs. Arrows indicate that no spores were produced at the lower limit of detection. Error bars represent standard deviations. b Northern blot showing the expression of Pxr-L and Pxr-S produced by the OC Δpxr-derived strains integrated with pxr homologs from different species of myxobacteria and the control strains. The asterisk marks binding of the probe to non-Pxr RNAs
In contrast, the Pxr paralogs in Cystobacter (Fig. 4a) varied greatly in their functional effects. Although both PxrCbm6.2 and PxrCbvi34.2 from C. minor Cbm 6 and C. violaceus Cbvi 34 effectively blocked spore production, PxrCb.1 and PxrCb.3 failed to do so. Spore production by the OC Δpxr-derived strains carrying these two paralogs was similar to that of OC Δpxr and the isogenic construct lacking any pxr sequence.
Structural differences in pxr paralogs affect levels of Pxr accumulation in M. xanthus
We used Northern analysis to examine Pxr-production patterns and levels for each pxr homolog (Fig. 4b). The three single-copy pxr homologs and pxr Cbm6.2 and pxr Cbvi34.2 from C. minor Cbm 6 and C. violaceus Cbvi 34 were all expressed into both Pxr-L and Pxr-S at sizes and levels similar to the native M. xanthus pxr allele, supporting the developmental phenotypes we observed from the strains carrying them. For the pxr Cb.1 and pxr Cb.3 paralogs that did not block development, Pxr-L was produced at a similar size as that from the M. xanthus allele but in much lower amounts. Pxr-S was not detected at all from the strain carrying pxr Cb.1 and was only barely detected in the strain carrying pxr Cb.3. These results indicate that lack of Pxr-S is associated with the inability of PxrCb.1 and PxrCb.3 to block development. This lack of Pxr-S may stem from the reduced amounts of Pxr-L, which in turn might be due to the instability of RNA transcripts.
If PxrCb.1 and PxrCb.3 transcripts are less stable than those of other alleles (at least when expressed in M. xanthus), such instability might be caused by the sequences per se or structural differences caused by their sequences. At the sequence level, the nucleotide changes that occurred in both pxr Cb.1 and pxr Cb.3 but not in pxr Cbm6.2 and pxr Cbvi34.2 are at positions 57, 81 and 87 (Fig. 1a). Although the nucleotide changes at positions 57 and 81 are different in pxr Cb.1 and pxr Cb.3, changes at these two positions might affect the stability of Pxr. The change at position 87 (T→C) is shared with pxr Mxs33 and pxr Mxs42, although it is possible that the interactions between this change and the ones at 57 and 81 also affect stability. The rest of the nucleotide differences in pxr Cb.1 and pxr Cb.3 not shared with pxr Cbm6.2 and pxr Cbvi34.2 do not overlap. Alternatively, these two distinct sets of differences might mediate similar changes (i.e. causing the instability of Pxr transcripts).
We calculated RNA structural stabilities with the self-folding free energy ΔG to explore the possibility that the sequences of pxr Cb.1 and pxr Cb.3 in Cystobacter affect the stability of their respective transcripts. None of the predicted structures from species other than M. xanthus (ΔG values from −58.9 to −52.2, with lower values reflecting greater stability) appear to be considerably less stable than the M. xanthus structure (ΔGGJV1 = −58.3). Therefore, it is unlikely that the reduced expression of PxrCb.1 and PxrCb.3 sRNAs in M. xanthus is caused by differences in intrinsic structural stabilities alone. Nevertheless, the re-computed free energies of PxrCb.1 and PxrCb.3 given the consensus structure are predicted to be substantially less stable (ΔGCb.1 = −38.9 and ΔGCb.3 = −15.2) than PxrCbm6.2, PxrCbvi34.2 and PxrGJV1 (ΔGCbm6.2 = −51.4, ΔGCbvi34.2 = −50.5 and ΔGGJV1 = −57.9), indicating that the base changes in pxr Cb.1 and pxr Cb.3 destabilize the consensus structure shared by single-copy Pxr orthologs.
Discussion
Bacterial sRNAs are a class of regulatory elements that has great importance in the function and evolution of bacterial genomes, but how their functions evolve over time remains poorly understood. Here, we tested for divergence of the bacterial sRNA Pxr in the fruiting myxobacteria by introducing sequences of its inferred ancestor and heterospecific homologs into an M. xanthus pxr deletion mutant. We showed that both the inferred ancestor and homologs from species with only one copy of pxr restored the regulatory function of Pxr observed in M. xanthus (Figs. 3-4), suggesting that such function has been established since its likely origin within the myxobacteria and has been conserved in different species (Fig. 5). Nevertheless, multiple paralogs present in the genus Cystobacter exhibited differences in their functional effects, with two paralogs controlling M. xanthus development but two others failing to do so (Figs. 4-5). These differences in functional effects may be due to differences in sRNA accumulation levels in turn caused by structural differences in the Pxr variants encoded by these paralogs. While previous work has examined functional differentiation of paralogous sRNAs in Vibrio spp. (e.g. [48–50]), here we have compared the functional effects of both orthologous and paralogous bacterial sRNAs in a common genetic background.
Fig. 5.
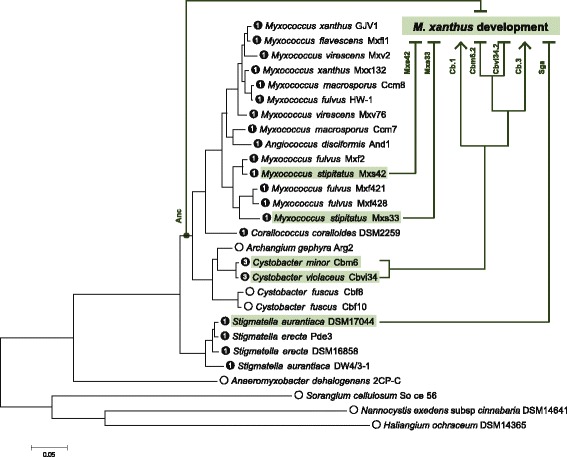
Phylogeny of myxobacterial species and strains summarizing the effects of diverse pxr alleles on M. xanthus development. The phylogenetic analysis was based on five conserved loci: 16S rRNA, 23S rRNA, pyrG, rpoB and pgm. Solid circles indicate that Pxr was detected in the respective species and the numbers inside indicate the numbers of pxr alleles found. Empty circles denote that Pxr was not detected. ‘Anc’ indicates the phylogenetic position of the ancestral pxr allele examined in this study. Shaded boxes highlight extant species from which respective pxr alleles were transferred into M. xanthus (see text for details). Lines towards the “M. xanthus development” box with bar heads and arrow heads indicate pxr alleles able vs. unable to control M. xanthus development, respectively. Modified from [38]
Our results also shed light on the sequence regions important for the function of Pxr. The pxr alleles we examined contain a specific set of polymorphic sites that allow us to infer their functional relevance. For the pxr ancestor and homologs that control development in M. xanthus, their sequences are highly identical in the first and part of the second stem-loops, illustrating the significance of these regions for the function of Pxr.
It has long been recognized by evolutionary biologists that genes from the same species coevolve together and during this process may become incompatible with genes from other species that are evolving independently due to reproductive isolation [51, 52]. Hybrid dysfunction caused by such independent divergence is well known in animals and plants [53]. Similarly, sRNAs from one species may not be able to control target mRNAs from another species, and this idea has been tested with the miR-310 miRNA family across different Drosophila species in animals [54]. For the sRNA Pxr in the myxobacteria, the homologs we introduced into M. xanthus are from species diversified in fruiting body size, shape and color [43]. However, many of their Pxr sRNAs controlled M. xanthus development in a manner similar to the native M. xanthus allele, revealing strong selective constraints on the regulatory interactions of Pxr (including both processing and interactions with target mRNAs) across different species. Our results thus suggest that Pxr targets are shared by these different species and will facilitate future identification of target genes. Nevertheless, our results did not exclude the possibility that regulons of Pxr have expanded in species other than M. xanthus, or the possibility that Pxr may evolve to regulate additional developmental processes in other species.
We also showed that the specificity of Pxr function appears to be evolving among some of its paralogs, as the sRNAs transcribed from pxr Cb.1 and pxr Cb.3 are expressed in much lower amounts in M. xanthus than are transcripts from the native M. xanthus allele and other homologs. Most likely, the structural differences caused by sequences in these two paralogs affect the stability of the sRNA molecules transcribed from them such that low accumulation of Pxr fails to effectively block development. In our experimental design, all Cystobacter paralogs are transcribed from the same M. xanthus enhancer binding site and promoter to eliminate potential confounding effects of any variation in the effects of these regulatory elements on transcription. Because all Cystobacter pxr paralogs have their own σ54 promoters, sequence differences in the enhancer binding sites and/or in the regulatory regions upstream of the promoters may mediate differential expression and enable higher expression of pxr Cb.1 and pxr Cb.3 in their native genomic background.
It is possible that Pxr interacts with accessory proteins that modulate their expression or stability. For example, the paralogous sequences or structures in Cystobacter might prevent or reduce processing of Pxr-L into Pxr-S by the relevant M. xanthus ribonuclease and thus prevent accumulation of the active form of Pxr. Nevertheless, our results show that even the Pxr-L forms from the Cystobacter alleles were present in much lower amounts. In E. coli, the RNA chaperone Hfq protects sRNAs from degradation by ribonucleases [33]. Nevertheless, Hfq preferentially binds to AU-rich sequences [55, 56] and no homologs of Hfq were detected in the sequenced myxobacterial genomes that are generally GC-rich, suggesting that an alternative protein might serve as RNA chaperone for Pxr sRNA. It is also known that in E. coli, polynucleotide phosphorylase (PNPase) increases the stability of sRNAs [57]. Such accessory proteins might identify specific sequence regions or structures of Pxr in order to regulate their expression or stability, and may have coevolved with Pxr closely in addition to target mRNAs. Identification of these proteins will allow testing for adverse interactions between Pxr sRNAs and accessory proteins from different species (e.g. Cystobacter species), and whether such interactions result in reduced accumulation levels of Pxr sRNAs we observed in this study. Lastly, it remains unknown whether these paralogous sRNAs effectively hybridize with respective target mRNA molecules in M. xanthus due to instability.
Known bacterial sRNA paralogs include OmrA and OmrB responsible for high osmolarity in E. coli [58], PrrF1 and PrrF2 for iron limitation in Pseudomonas aeruginosa [39], four copies of Qrr for quorum sensing in Vibrio cholerae [59] and five copies in the bioluminescent marine V. harveyi [50] (for detailed review, see [60]). Interestingly, while the Qrr sRNAs in V. cholerae act redundantly, in V. harveyi they act additively to control the quorum sensing master regulator [50, 59], and some of the sRNAs have evolved preferences for additional targets [48, 49]. Our findings here add another example of divergence among bacterial sRNA paralogs, and lay the groundwork to study how developmental programs are regulated by sRNAs in myxobacterial species. Future work of interest includes eludication of the effects of each pxr paralog in their own host species in the Cystobacter subclade.
Conclusions
In animals and plants, the discovery of RNA silencing led to the understanding that small non-coding RNAs are commonly used by eukaryotes to post-transcriptionally regulate the expression of many genes. In bacteria, a rapidly increasing number of small RNAs have also been found to regulate a wide variety of important biological processes. However, in most systems the functional evolution of sRNAs and how they coevolve with their associated regulatory networks remain poorly understood. Pxr is a small RNA that regulates the initiation of fruiting body development in the model myxobacterial species M. xanthus and appears to have a recent origin in a subclade of the myxobacteria, thus serving as a great system to study the divergence of a bacterial sRNA. We show that the regulatory function of Pxr is conserved across different myxobacterial species, generating insights into identifying functional elements and the sequence-function relations of bacterial sRNAs. Further, its function appears to have changed in some lineages through gene duplication and subsequent diversification. This provides opportunities to study the roles of sRNAs in developmental programs of the myxobacteria, which exhibit among the most complex multicellular behaviors found in bacteria.
Methods
Plasmid construction
All plasmids constructed in this study are listed in Table 1. One positive and one negative control plasmids (pPxrGJV1 and pPxrnull, respectively) were first constructed. To construct the positive control pPxrGJV1 that bears the GJV1 pxr allele, a 537-bp PCR fragment containing the 3′-terminal 167-nt of nla19, the 262-nt intergenic region between nla19 and pxr that contains the predicted σ54 promoter, and the 108-nt pxr coding region in M. xanthus was cloned into pCR2.1 vector that carries a kanamycin-resistance marker gene. To construct the negative control pPxrnull that carries only the 429-nt region upstream of the pxr gene, a PCR fragment containing only the 3′-terminal 167-nt of nla19 and the 262-nt intergenic region between nla19 and pxr was cloned into pCR2.1. Primers used for PCR amplification are listed in Additional file 1: Table S1. Competent cells of Escherichia coli TOP10 were used for plasmid cloning and were grown at 37 °C in Luria-Bertani (LB) medium [61] or on X-Gal/IPTG LB hard (1.5%) agar plates to screen for colonies with PCR fragment inserts. The sequences and orientation of the inserts were confirmed by sequencing.
The inferred pxr ancestor
To reconstruct the previously inferred pxr ancestor [38] in the laboratory, we used the genomic DNA of M. xanthus GJV1 as template and designed three PCR primers that contain the ancestral nucleotides in the pxr ancestor in order to replace the M. xanthus-specific sequence. Two of the primers (Node38_Bulge_For and Node38_Bulge_Rev) are the sense and anti-sense strand sequences, respectively, at the ancestral pxr positions 13–33 and contain the ancestral U on the bulge on the left side of the first stem-loop (Fig. 1a, b). The third primer, Node38_LongRev, is a 64-nt long primer at the positions 48–111 that contains the remaining six ancestral sites from the third stem-loop to the end of the pxr coding region. We first PCR-amplified a 462-bp DNA fragment that contains the 429-bp fragment preceding pxr in conjunction with the nucleotide positions 1–33 in the ancestral pxr with the primers GV367 and Node38_Bulge_Rev (5′ GGG GGG AAC CAC CTT CAG CCT 3′) (the bold letter indicates the ancestral nucleotide). Next, we PCR-amplified a 99-bp fragment from the positions 13 to 111 in the pxr ancestor using primers Node38_Bulge_For (5′ AGG CTG AAG GTG GTT CCC CCC 3′) and Node38_LongRev (5′ AAA AGA AGG CGG CCC GAT ACC CCA AGA GAG GGT ACC GGG CCG CGG GTT CTT CTA AAG GTG ACT C 5′). We subsequently performed a PCR to join the 462-bp and 99-bp fragments together by the overlapping sequences at the positions 13–33 in the pxr ancestor and to amplify the joint 540-nt fragment with GV367 and Node38_LongRev. The resulting PCR fragments were cloned into pCR2.1 and verified by sequencing.
Homologs of pxr from different myxobacterial species
To introduce different pxr homologs into M. xanthus, we first PCR-amplified the 429-bp DNA fragment preceding pxr coding region in M. xanthus. We then PCR-amplified each pxr homolog using the genomic DNA extracted from their host strains with primer sequences listed in Additional file 1: Table S1. Next, we ligated the 429-bp PCR products with each pxr homolog using T4 DNA ligase (Thermo Scientific). A subsequent PCR was performed to amplify the ligated fragment using the forward primer complementary to the 5′ region of the 429-bp fragment and the reverse primer complementary to the 3′ region of each pxr homolog. The resulting PCR products were cloned into pCR2.1 and confirmed by sequencing.
M. xanthus strain construction
All M. xanthus strains used in this study are listed in Table 1. All constructed strains were derived from the strain OC Δpxr and generated with plasmid integration, which was achieved by electroporation of OC Δpxr competent cells with plasmids that carry the 429-nt homologous region with the predicted σ54 promoter preceding pxr in conjunction with different versions of pxr. To perform electroporation, overnight cultures of OC Δpxr grown in CTT liquid medium [62] were harvested at exponential-growth phase by centrifugation at room temperature. The cells were washed five times with and resuspended in double-distilled water. Seventy-five microliters of the cell suspension and 3 μl of the plasmid DNA were mixed and transferred to electroporation cuvette. Electroporation was performed with a Bio-Rad Gene Pulser apparatus set at 400 Ω, 25 μF and 0.65 kV. The cells were then transferred to an Erlenmeyer flask containing 3 ml of CTT liquid and incubated overnight at 32 °C with constant shaking at 300 rpm. Samples were diluted into CTT soft (0.5%) agar containing 40 μg kanamycin/ml and incubated at 32 °C with 90% rH (relative humidity) for a week until colonies became visible. For each constructed strain, two to three clones were isolated. The genetic organization of the resulting strains is illustrated in Fig. 2.
Developmental assay
M. xanthus strains were grown in CTT liquid at 32 °C with constant shaking at 300 rpm. To perform the developmental assays, cultures in exponential-growth phase were centrifuged at room temperature and resuspended in TPM liquid [63] to a density of 5 × 109 cells per ml. For each strain, 50 μl of resuspended cells were spotted on the center of nutrient free TPM hard (1.5%) agar plates and incubated at 32 °C with 90% rH. After three days, cells were harvested with a scalpel blade, transferred into 1 ml of double-distilled water and heated at 50 °C for two hours to select for heat-resistant spores. Samples were then sonicated with a microtip to disperse spores, diluted and plated into CTT soft (0.5%) agar plates. After seven days, visible colonies were counted to estimate the total number of spores produced during development. In cases where no colonies grew at our lowest dilution factor (10−1), a value of ten spores per replicate was entered for data analysis, providing conservatively high estimates of spore production in these cases. In any given assay, two or three clones of identical constructs were tested. The same experimental procedure was repeated at least three times for all strains.
Expression pattern and stability of Pxr sRNA
We detected the expression of Pxr sRNA by Northern blotting. The total enriched small-sized RNA preparation of each strain was extracted from vegetative cultures growing in CTT liquid with the mirVana miRNA isolation kit (Ambion). RNA concentration was measured with a Nanodrop spectrophotometer and equal amounts of RNA (1 μg) were electrophoresed in 10% SequaGel (National Diagnostics) and electro-transferred onto a BrightStar®-Plus positively charged nylon membrane (Ambion). After UV cross-linking, the membrane was pre-hybridized in 4 ml UltraHyb-Oligo buffer for 30 min and subsequently hybridized in the same solution containing 100 pmol 3’Biotin-TEG-pxr oligo probe (5′-ACC GGA AGT GCT GAA GGG GTG GGG GG-3′) (Sigma) overnight. Pxr sRNA was detected with BrightStar® Biodetect non-isotopic kit (Ambion). We estimated the stabilities of the Pxr ancestor and homologs in one stem-loop structure by computing the self-folding free energy ΔG of their RNA sequences using Mfold [64]. The consensus secondary structure of Pxr based on the alignment of single-copy pxr homologs was predicted with RNAalifold within the ViennaRNA software package [65]. We re-estimated the ΔG of the homologs given the consensus structure with the RNAeval function [66].
Acknowledgements
The authors thank anonymous reviewers for comments on the manuscript and members of the Velicer lab for their support during the study.
Availability for data and materials
The strains generated during the study are available from the corresponding author on reasonable request. All data generated or analyzed during the study are included in this published article.
Funding
This work was supported by U.S. National Institutes of Health Grant R01 GM079690 and Indiana University (Bloomington, IN, USA).
Abbreviations
- miRNAs
microRNAs
- Pxr-L
the long form of Pxr sRNA
- Pxr-S
the short form of Pxr sRNA
- sRNAs
small non-coding RNAs
Additional file
Primers used in this study. (DOCX 17 kb)
Authors’ contributions
ICKC, GJV and YTNY designed research; ICKC and YTNY performed research; ICKC analyzed data; ICKC, GJV and YTNT wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-017-1037-5) contains supplementary material, which is available to authorized users.
Contributor Information
I-Chen Kimberly Chen, Email: kichen.evol@gmail.com.
Gregory J. Velicer, Email: gregory.velicer@env.ethz.ch
Yuen-Tsu Nicco Yu, Email: nicco.yu@env.ethz.ch.
References
- 1.Babski J, Maier LK, Heyer R, Jaschinski K, Prasse D, Jager D, Randau L, Schmitz RA, Marchfelder A, Soppa J. Small regulatory RNAs in Archaea. RNA Biol. 2014;11(5):484–493. doi: 10.4161/rna.28452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding Frontiers. Mol Cell. 2011;43(6):880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10(9):632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 6.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 7.Yu YT, Yuan X, Velicer GJ. Adaptive evolution of an sRNA that controls Myxococcus development. Science. 2010;328(5981):993. doi: 10.1126/science.1187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masse E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10(2):140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Moller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16(13):1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boysen A, Moller-Jensen J, Kallipolitis B, Valentin-Hansen P, Overgaard M. Translational regulation of gene expression by an anaerobically induced small non-coding RNA in Escherichia coli. J Biol Chem. 2010;285(14):10690–10702. doi: 10.1074/jbc.M109.089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durand S, Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol. 2010;75(5):1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20(17):2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- 13.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol. 2006;9(6):605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 14.De Lay N, Gottesman S. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol. 2009;191(2):461–476. doi: 10.1128/JB.01157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overgaard M, Johansen J, Moller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Mol Microbiol. 2009;73(5):790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- 16.Bardill JP, Hammer BK. Non-coding sRNAs regulate virulence in the bacterial pathogen Vibrio cholerae. RNA Biol. 2012;9(4):392–401. doi: 10.4161/rna.19975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers JR, Sauer K. Small RNAs and their role in biofilm formation. Trends Microbiol. 2013;21(1):39–49. doi: 10.1016/j.tim.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bejerano-Sagie M, Xavier KB. The role of small RNAs in quorum sensing. Curr Opin Microbiol. 2007;10(2):189–198. doi: 10.1016/j.mib.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37(4):856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 20.Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol. 2004;54(4):1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 21.Horler RS, Vanderpool CK. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res. 2009;37(16):5465–5476. doi: 10.1093/nar/gkp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21(21):2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao Y, Zhang ZJ, Erickson DW, Huang M, Huang Y, Li J, Hwa T, Shi H. Quantifying the sequence-function relation in gene silencing by bacterial small RNAs. Proc Natl Acad Sci U S A. 2011;108(30):12473–12478. doi: 10.1073/pnas.1100432108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol Microbiol. 2006;61(4):1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 25.Papenfort K, Podkaminski D, Hinton JC, Vogel J. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc Natl Acad Sci U S A. 2012;109(13):E757–E764. doi: 10.1073/pnas.1119414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterman N, Lavi-Itzkovitz A, Levine E. Large-scale mapping of sequence-function relations in small regulatory RNAs reveals plasticity and modularity. Nucleic Acids Res. 2014;42(19):12177–12188. doi: 10.1093/nar/gku863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9(8):578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey SP, Minesinger BK, Kumar J, Walker GC. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 2011;39(11):4691–4708. doi: 10.1093/nar/gkr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, Helmann JD. The Bacillus subtilis Iron-sparing response is mediated by a fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A. 2008;105(33):11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol. 2009;74(1):139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 31.Davis BM, Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol Microbiol. 2007;65(2):373–385. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11(12):941–950. doi: 10.1016/S0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 33.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17(19):2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viegas SC, Silva IJ, Saramago M, Domingues S, Arraiano CM. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res. 2011;39(7):2918–2930. doi: 10.1093/nar/gkq1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol. 2009;16(8):840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 36.Peer A, Margalit H. Evolutionary patterns of Escherichia coli small RNAs and their regulatory interactions. RNA. 2014;20(7):994–1003. doi: 10.1261/rna.043133.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toffano-Nioche C, Nguyen AN, Kuchly C, Ott A, Gautheret D, Bouloc P, Jacq A. Transcriptomic profiling of the oyster pathogen Vibrio splendidus opens a window on the evolutionary dynamics of the small RNA repertoire in the Vibrio genus. RNA. 2012;18(12):2201–2219. doi: 10.1261/rna.033324.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen IC, Griesenauer B, Yu YT, Velicer GJ. A recent evolutionary origin of a bacterial small RNA that controls multicellular fruiting body development. Mol Phylogenet Evol. 2014;73:1–9. doi: 10.1016/j.ympev.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A. 2004;101(26):9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skippington E, Ragan MA. Evolutionary dynamics of small RNAs in 27 Escherichia coli and Shigella genomes. Genome Biol Evol. 2012;4(3):330–345. doi: 10.1093/gbe/evs001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wadler CS, Vanderpool CK. Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res. 2009;37(16):5477–5485. doi: 10.1093/nar/gkp591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci U S A. 2007;104(51):20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawid W. Biology and global distribution of myxobacteria in soils. FEMS Microbiol Rev. 2000;24(4):403–427. doi: 10.1111/j.1574-6976.2000.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 44.Fiegna F, Yu YT, Kadam SV, Velicer GJ. Evolution of an obligate social cheater to a superior cooperator. Nature. 2006;441(7091):310–314. doi: 10.1038/nature04677. [DOI] [PubMed] [Google Scholar]
- 45.Velicer GJ, Raddatz G, Keller H, Deiss S, Lanz C, Dinkelacker I, Schuster SC. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc Natl Acad Sci U S A. 2006;103(21):8107–8112. doi: 10.1073/pnas.0510740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Velicer GJ, Kroos L, Lenski RE. Loss of social behaviors by Myxococcus xanthus during evolution in an unstructured habitat. Proc Natl Acad Sci U S A. 1998;95(21):12376–12380. doi: 10.1073/pnas.95.21.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci U S A. 2006;103(41):15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao Y, Feng L, Rutherford ST, Papenfort K, Bassler BL. Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. EMBO J. 2013;32(15):2158–2171. doi: 10.1038/emboj.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao Y, Bassler BL. Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol Microbiol. 2012;83(3):599–611. doi: 10.1111/j.1365-2958.2011.07959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21(2):221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobzhansky T. Further data on the variation of the Y chromosome in drosophila Pseudoobscura. Genetics. 1937;22(3):340–346. doi: 10.1093/genetics/22.3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muller HJ. Isolating mechanisms, evolution and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- 53.Coyne JA, Orr HA. Speciation. Sinauer: Sunderland, Massachusetts; 2004. [Google Scholar]
- 54.Tang T, Kumar S, Shen Y, Lu J, Wu ML, Shi S, Li WH, Wu CI. Adverse interactions between micro-RNAs and target genes from different species. Proc Natl Acad Sci U S A. 2010;107(29):12935–12940. doi: 10.1073/pnas.1007591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10(2):125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 56.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51(6):1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 57.De Lay N, Gottesman S. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA. 2011;17(6):1172–1189. doi: 10.1261/rna.2531211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59(1):231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 59.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118(1):69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 60.Caswell CC, Oglesby-Sherrouse AG, Murphy ER. Sibling rivalry: related bacterial small RNAs and their redundant and non-redundant roles. Front Cell Infect Microbiol. 2014;4:151. doi: 10.3389/fcimb.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 62.Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A. 1977;74(7):2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bretscher AP, Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978;133(2):763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]


