Abstract
Background
Ciliated muconodular papillary tumors (CMPTs) are newly recognized rare peripheral lung nodules that are histologically characterized by ciliated columnar, goblet, and basal cells. Although recent studies have shown that CMPTs constitute a neoplastic disease, the complete histogenesis of CMPTs is not fully understood and molecular data are limited.
Methods
We reviewed four cases of CMPT and performed immunohistochemical and genomic analyses to establish CMPT profiles.
Results
All cases were positive for hepatocyte nuclear factor-4α and mucin 5B and negative for programmed death ligand 1 expression, as determined by immunohistochemistry. The genetic analysis revealed three pathogenic mutations (BRAF V600E, AKT1 E17K, and KRAS G12D), with the KRAS mutation reported here for the first time.
Conclusion
Histological and genetic profiles indicate that CMPTs are likely neoplastic and exhibit features similar to mucinous adenocarcinoma. This suggests that some CMPTs may be a precursor lesion of mucinous adenocarcinoma.
Electronic supplementary material
The online version of this article (doi:10.1186/s13000-017-0651-2) contains supplementary material, which is available to authorized users.
Keywords: Ciliated muconodular papillary tumor, CMPT, Next-generation sequencing, Mutation, BRAF, RAS, AKT1
Background
Ciliated muconodular papillary tumors (CMPTs) are a newly recognized small-size papillary tumor of the peripheral lung that contain columnar cells, occasional basal cells, and mucus-producing cells as well as extracellular mucin pools of various sizes. Although CMPTs were first described based on certain pathological features that suggested a malignant potential, similar diseases such as extremely well-differentiated papillary adenocarcinoma with prominent cilia formation have been reported under different names [1–3]. Due to its complex histology and presence of inflammation and fibrosis, the metaplastic nature of CMPTs has been debated. However, recent reports have revealed the frequent presence of driver gene mutations, and CMPT is now recognized as a neoplasia [4–6]. The histogenesis and molecular characteristics of CMPTs are not well understood owing to the rarity of the disease. To address this issue, we reviewed our case archive and characterized CMPTs by immunohistochemistry and next-generation sequencing (NGS).
Methods
Patient selection and tissue preparation
Cases of surgically resected CMPT from 2012 and 2016 were searched in the pathology archive of Nagasaki University Hospital. Hematoxylin and eosin-stained slides of each case were reviewed by two pathologists specializing in thoracic medicine. We identified four CMPT cases; two of these had been originally diagnosed as CMPT, whereas the other cases had been diagnosed as glandular papilloma and mucinous adenocarcinoma. Clinical data were extracted from the hospital’s electronic medical records. Sections with a thickness of 4 μm from formalin-fixed, paraffin-embedded (FFPE) tissue samples were examined by immunohistochemistry, and 15 sections with a thickness of 5 μm were subjected to NGS of 50 cancer-related genes using the Ion Torrent PGM system (Life Technologies, Carlsbad, CA), and10 μm thick section on the conventional glass slide were submitted for 29 genes analysis using MiSeq (Illumina, San Diego, CA, USA).
Isolation of genomic DNA
Genomic DNA was extracted from FFPE tumor samples using the QIAamp DNA FFPE kit (Qiagen, Valencia, CA, USA), and the concentration was determined using the Quant-iT PicoGreen dsDNA Assay kit (Life Technologies).
Targeted deep sequencing of mutational hotspots in 50 cancer-related genes
Genomic DNA was subjected to whole-exome sequencing using the Ion AmpliSeq Cancer Hotspot Panel v.2 (Life Technologies) according to the manufacturer’s instructions. The purified library was sequenced on an Ion PGM instrument using Ion PGM Hi-Q Sequencing kit and Ion 318 Chip Kit v.2 (all from Life Technologies). DNA sequencing data were accessed with the Torrent Suite v.5.0 program (Life Technologies). The coverage analysis was performed using the coverage analysis plug-in v5.0. Reads were aligned with the hg19 human reference genome, and potential mutations were identified using Variant Call Format v.5.0. Raw variant calls were annotated with CLC Genomics Workbench software (CLC bio, Aarhus, Denmark). Variants were manually verified using the integrative genomics viewer (Broad Institute, Cambridge, MA, USA). Known single nucleotide polymorphisms were identified using the Human Genetic Variation Database (http://www.hgvd.genome.med.kyoto-u.ac.jp/) [7] and were excluded.
We also performed sequencing on Illumina MiSeq instrument. As recommended by the manufacturer, 50 ng of dsDNA was used for generating sequencing libraries using Find-It cancer hotspot panel (Contextual Genomics, Vancouver, BC, Canada). The panel targets 90+ cancer hotspot mutations including eight coding exons in 29 cancer-related genes. DNA libraries were denatured and diluted as per Illumina’s recommendations. Samples were run on MiSeq machine using 300 cycle MiSeq Reagent Kit V2 (Cat.No.MS-102-2002, Illumina).
Immunohistochemistry
Immunohistochemical analysis of 4-μm tissue sections was carried out using the Ventana Bench Mark XT Automated stainer (Ventana Medical Systems, Tucson, AZ, USA) and BOND III fully automated stainer (Leica Biosystems, Melbourne, Australia) with antibodies against thyroid transcription factor (TTF)-1 (clone SPT24; Leica Novocastra, Newcastle Upon Tyne, UK), hepatocyte nuclear factor (HNF)-4α (clone H1415; Perseus Proteomics, Tokyo, Japan), mucin (MUC)5B (Santa Cruz Biotechnology, Santa Cruz, CA, USA), MUC5A (clone CLH2; Leica Novocastra), cytokeratin (CK)7 (clone OV-TL12/30; Dako Japan, Tokyo, Japan), CK20 (clone Ks20.8; Dako Japan), p53 (clone DO-7; Leica Novocastra), Ki-67 (clone MIB-1; Dako Japan), Caudal-type homeobox (CDX)2 (clone EPR2764Y; Roche Diagnostics, Indianapolis, IN, USA), ALK (clone D5F3; Roche Diagnostics), BRAF V600E (clone VE1; Roche Diagnostics) and programmed death ligand (PD-L)1 (clone SP142; Spring Bioscience, Pleasanton, CA, USA). Details of optimized staining protocols are listed in Additional file 1. HNF4α, MUC5B, MUC5A, CK7, and CK20 staining was scored as follows: −, negative; 1+, focal; and 2+, diffuse. The intensity of p53 expression was assessed as follows: 1+, faint and sporadic; 2+, frequently positive with no or equivocal overexpression; and 3+, unequivocal and diffuse overexpression. BRAF V600E and ALK expression was judged as positive when strong and diffuse staining was observed in cancer cells. The percentage of Ki-67-immunopositive cells was analyzed using a nuclear algorithm (Nuclear_v.9_v.10.0.0.1798; Imagescope, Leica Biosystems, Nussloch, Germany). PD-L1 expression was scored as the percentage of PD-L1-positive cells among tumor cells. CDX2 expression was scored as follows: −, negative; 1+, ≤ 25% positive; 2+, > 25 and <50% positive; and 3+, ≥ 50% positive. Two observers (E.U. and J.F.) independently scored the staining and a consensus was obtained through discussion when there was a discrepancy.
Results
Clinical summary
Clinical data for the four CMPT patients are summarized in Table 1. The patients were all female with a median age of 67 years old. None had a history of smoking. Tumors ranged in size from 8 to 25 mm (median: 11 mm). Lobectomy was performed in three cases, and segmentectomy was carried out in one case.
Table 1.
Summary of clinical data and detected gene mutations in the past reports and present cases
| Author | Age (median) | Sex (M/F) | smoking habit (Y/N) | Driver Mutation | Other Mutations | NGS panel |
|---|---|---|---|---|---|---|
| Kamata et al. [4] | 62 | 7/3 | 5/5 | BRAF V600E, 4 cases; EGFR ex19del E746-T751, 3 cases; BRAF G606R, 1 case | IDH1 G123R, 1 case; CTNNB1 D32N, 1 case; PTPN11 E76K, 1 case; PTPN11 P491L, 1 case; TP53 L289F, 1 case | Ion AmpliSeq Cancer Hotspot Panel v2 |
| Liu et al. [5] | – | 1/3 | – | BRAF V600E, 1 case | AKT1 E17K, 1 case | Ion AmpliSeq Cancer Hotspot Panel v2 |
| Lau et al. [15] | 19 | 0/1 | 0/1 | none | none | Ion Ampliseq Colon and lung cancer panel |
| Our series | 67 | 0/4 | 0/4 | BRAF V600E 1 caseKRAS G12D, 1 case | AKT1 E17K, 1 case | Ion AmpliSeq Cancer Hotspot Panel v2; The Contextual Genomics Find-ItTM test |
F female, M male
Histological findings
The histological features of the four CMPT cases were consistent with those previously reported [1, 8, 9]. That is, the tumors typically exhibited a mixture of acinar and papillary growth patterns without clear evidence of invasion, and were surrounded by abundant mucus pools in the alveolar spaces (Fig. 1a). The tumors were composed of two basal cell layers and surface epithelia. The latter consisted of an uneven mosaic of ciliated columnar, goblet, and mucin-producing epithelial cells of the gastric type (Fig. 1b). Few tumor cells showed nuclear atypia, and no mitosis or necrosis was observed.
Fig. 1.
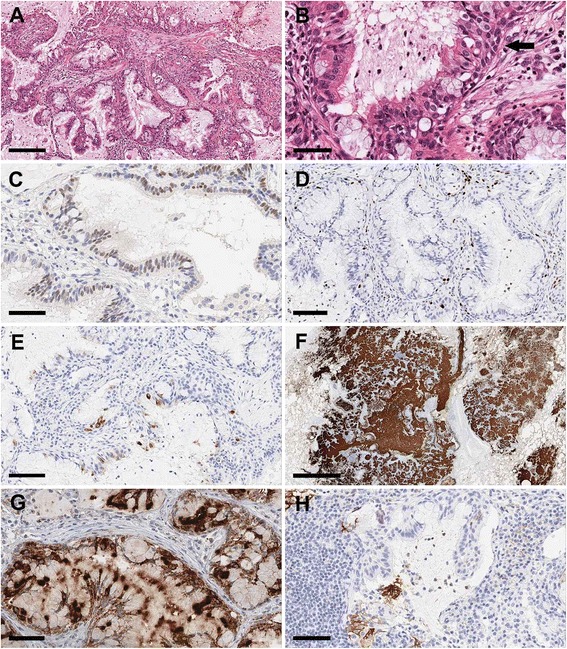
Immunohistochemical analysis of CMPTs. a Low magnification view of a CMPT showing papillary epithelial proliferation with abundant mucus production. b Higher magnification view of the same case showing a mixture of goblet, ciliated columnar, and basal cells (arrow). c HNF-4α positivity in epithelial cell nuclei. Scale bar = 60 μm. d The positive rate for Ki-67 expression was <10%. Scale bar = 100 μm. e Focal MUC5AC staining in occasional ciliated cells. Scale bar = 90 μm. f Epithelial cells and mucin were strongly positive for MUC5B. Scale bar = 2 mm. g BRAF V600E staining was strong and diffuse in most epithelial cells of a case harboring BRAF V600E and AKT1 E17K mutations. Scale bar = 60 μm. h PD-L1 staining was mostly negative (< 1%); however, focal membranous immunoreactivity was observed. Scale bar = 70 μm
Immunohistochemistry
All four cases were positive for nuclear HNF4α and TTF-1. Two cases showed diffuse and strong staining for HNF4α, as seen in mucinous adenocarcinoma or colorectal carcinoma (Fig. 1c). The Ki-67 index was low (2.5%–10%), consistent with a less aggressive nature (Fig. 1d). In contrast to mucinous adenocarcinoma, all cases examined in this study showed diffuse TTF-1 positivity and one case showed possible overexpression. There was sparse cytoplasmic expression of MUC5AC (Fig. 1e) while diffuse MUC5B—which is very rare in the normal lung parenchyma—was detected in all cases (Fig. 1f). Nuclear expression of p53 was sporadic in one case and more frequent in two cases, and in one case p53 was clearly overexpressed. There were no or only a few cells that were positive for CDX2 and CK20. p63 expression highlighted the basal layers in all cases, although the area of coverage was focal in two cases and broad in the two others (Table 2).
Table 2.
Summary of immnohistochemical findings in previouse reports and present cases
| Author | TTF-1 | Ki67 | CK7 | CK20 | MUC5AC | MUC5B | p53 | HNF4α | p63/p40 | CDX2 | PD-L1 | ALK | BRAF V600E |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sato et al. [8] | 2/2 | 3%; 10% | 2/2 | 0/2 | 1/2 | - | - | - | - | - | - | - | - |
| Chuang et al. [14] | 1/1 | <1% | 1/1 | 0/1 | - | - | 0/1 | - | 1/1 | - | - | - | - |
| Chu et al. [13] | 1/1 | - | 1/1 | 0/1 | - | - | - | - | 1/1 | - | - | - | - |
| Kon et al. [12] | 5/5 | <1%; 5 | 5/5 | 0/5 | 0/5 | - | 0/5 | - | 5/5 | - | - | - | - |
| Lau et al. [15] | 1/1 | not increase | 1/1 | 0/1 | 1/1(weak) | - | - | - | 1/1 | - | - | - | - |
| Kamata et al. [4, 11] | +a | - | - | - | +a (cliated cells) | - | - | 0/10a | +a | - | - | - | 4/4b |
| Ishikawa et al. [16] | 3/5 | <5% 3; <10% 1 | 5/5 | 0/5 | - | - | 3/3 | - | - | - | - | - | - |
| Taguchi et al. [10] | 0/1 | 3.7% | 1/1 | 0/1 | 1/1 | - | <1% | - | 1/1 | - | - | 1/1 | 0/1 |
| Jin et al. [6] | 1/1 | - | 1/1 | - | - | - | - | - | 1/1 | - | - | 1/1 | - |
| Our series | 4/4 | <5% 3; <10% 1 | 4/4 | 0/4 | 4/4 | 4/4 | 1/4 | 4/4 | 4/4d | 0/4 | <1% 4/4 | 0/4 | 1/4c |
-; not available, anumbers of positive/negative not reported, bcases with BRAF mutation, ccase with BRAF mutation was positive (1/1), done case was a few as positive
One case with BRAF V600E mutation identified by NGS showed diffuse and strong BRAF V600E staining, while the other three cases showed none (Fig. 1g). There was no ALK and EGFR mutations identified in our cases. ALK status was also confirmed by immunohistochemistry.
All cases exhibited <1% PD-L1 positivity in tumor cell membranes (Fig. 1h). These findings suggest that CMPTs are tumors that originated from a terminal respiratory unit and are showed features of gastric-type glands.
Genetic analysis
Gene mutations were detected in two of the four cases (50%) (Table 1). One case harbored BRAF V600E and AKT1 E17K mutations—which was interestingly identical as what have been recently reported [4, 5]—and another had a KRAS G12D mutation. The detected mutation status was identical on both Ion PGM instrument system and Illumina MiSeq system. A KRAS G12C mutation was found in one of the two other cases; however, the significance of this mutation is unclear due to its low frequency.
Discussion
We carried out immunohistochemical and molecular analyses of four cases of CMPT and identified a previously unreported KRAS mutation in addition to known BRAF and AKT1 mutations. The findings of BRAF and AKT1 mutations were identical to those reported in recent studies [4, 5]. However, we could not identify either EGFR or ALK mutation in our cases [4, 6, 10].
Interestingly, the case with a KRAS G12D mutation detected by Ion PGM and Illumina method showed fewer basal cell layers, as confirmed by p63 immunostaining, while another case with reduced basal cell layer coverage also had a KRAS mutation (G12C). We did not take this mutation into account due to its low frequency by Ion PGM method and negative result by Illumina method. This KRAS status could be due to intratumoral heterogeneity—i.e., the few cells harboring this mutation may have been overshadowed by wild-type cells, which constituted the majority of the tumor cells.
Our immunostaining results were consistent with previous reports [6, 8, 10–16]. However, we showed for the first time that CMPTs were positive for HNF-4α and MUC5B. Although these tumors show similarities to mucinous adenocarcinoma, there are significant differences between them such as the presence of cilia in columnar epithelia and basal cell intervention in the latter. In addition, invasive mucinous adenocarcinoma is usually TTF-1-negative and has distinct malignant features [17].
Conclusion
This is the first report of CMPTs of the lung harboring a KRAS mutation. Our findings suggest that CMPT cases are heterogeneous, and some CMPT cases may be a precancerous form of invasive mucinous adenocarcinoma, although additional studies are needed to investigate this possibility.
Acknowledgments
The authors are sincerely grateful to Professor Hiroaki Ikeda, Department of Oncology, Nagasaki University Graduate School of Biomedical Sciences, Professor Kazuto Ashizawa, Unit of Translational Medicine, Department of Clinical Oncology, Nagasaki University Graduate School of Biomedical Sciences, Assistant Professor Hiroyuki Yamaguchi, Second Department of Internal Medicine, Nagasaki University School of Medicine, Professor Mitsuko Masutani, Department of Frontier Life Sciences, Nagasaki University Graduate School of Biochemical Sciences, Associate professor Chizu Fukushima, Clinical Research Center, Nagasaki University Hospital for supporting NGS research by the Nagasaki University Priority Research Subject Project Based on Medium-term Goals and Plans fund.
The authors are sincerely grateful to Masakazu Souda, Department of Pathology, Nagasaki University Graduate School of Biomedical Sciences, Yuki Imaoka, Department of Pathology, Nagasaki University Graduate School of Biomedical Sciences for their technical assistances.
Funding
Nagasaki University Priority Research Subject Project Based on Medium-term Goals and Plans.
Availability of data and materials
All data is available upon request to the corresponding author.
Abbreviations
- (HNF)-4α
Hepatocyte nuclear factor 4α
- ALK
Anaplastic lymphoma kinase
- CDX2
Caudal-type homeobox 2
- CK
Cytokeratin
- CMPT
Ciliated muconodular papillary tumor
- EGFR
Epidermal growth factor receptor
- FFPE
Formalin-fixed, paraffin-embedded
- MUC
Mucin
- NGS
Next generation sequencing analysis
- PD-L1
Programmed death ligand 1
- TTF-1
Thyroid transcription factor-1
Additional file
Antibodies used for immunohistochemistry. (XLSX 10 kb)
Authors’ contributions
EU conceived of the study design, participated in immunostaining, molecular analysis of NGS and execution of the study. BF received a research grant, conceived of the study design and reviewed manuscript. KS supervised all procedure of NGS and assisted in drafting the manuscript. PL supervised all procedure of NGS and assisted in drafting the manuscript. TT participated in histological analysis. YK participated in providing clinical data and execution of the study. TT participated in providing clinical data. NY participated in providing clinical data. TN participated in providing clinical data. KN participated in molecular analysis, supervised NGS and execution of the study. JF conceived of the study design, and reviewed manuscript. All authors participated for data analysis and approved the final version.
Ethics approval and consent to participate
All procedures performed in this study were approved by the ethical committee at Nagasaki University (approval no.16052328).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13000-017-0651-2) contains supplementary material, which is available to authorized users.
Contributor Information
Emiko Udo, Email: udo-e@nagasaki-u.ac.jp.
Bungo Furusato, Phone: +81-95-819-7055, Email: bfurusato@nagasaki-u.ac.jp.
Kazuko Sakai, Email: kasakai@med.kindai.ac.jp.
Leah M Prentice, Email: Lprentice@contextualgenomics.com.
Tomonori Tanaka, Email: tanaka.t.tomonori@gmail.com.
Yuka Kitamura, Email: ykita@apricot.ocn.ne.jp.
Tomoshi Tsuchiya, Email: tomoshi@nagasaki-u.ac.jp.
Naoya Yamasaki, Email: ynaoya@nagasaki-u.ac.jp.
Takeshi Nagayasu, Email: nagayasu@nagasaki-u.ac.jp.
Kazuto Nishio, Email: knishio@med.kindai.ac.jp.
Junya Fukuoka, Email: fukuokaj@nagasaki-u.ac.jp.
References
- 1.Ishikawa Y. Ciliated muconodular papillary tumor of the peripheral lung: benign or malignant. Patholo Clin Med (Bryouri-to-Rinsho) 2002;20:964–965. [Google Scholar]
- 2.Nakamura S, Koshikawa T, Sato T, Hayashi K, Suchi T. Extremely well differentiated papillary adenocarcinoma of the lung with prominent cilia formation. Acta Pathol Jpn. 1992;42:745–750. doi: 10.1111/j.1440-1827.1992.tb03225.x. [DOI] [PubMed] [Google Scholar]
- 3.Harada T, Akiyama Y, Ogasawara H, Kishi F, Hattori A, Okamoto K, Hiramatsu M, Ishikawa Y. Ciliated muconodular papillary tumor of the peripheral lung: a newly defined rare tumor. Respir Med. 2008;1:3. [Google Scholar]
- 4.Kamata T, Sunami K, Yoshida A, Shiraishi K, Furuta K, Shimada Y, Katai H, Watanabe S, Asamura H, Kohno T, Tsuta K. Frequent BRAF or EGFR mutations in ciliated muconodular papillary tumors of the lung. J Thorac Oncol. 2016;11:261–265. doi: 10.1016/j.jtho.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, Aesif SW, Kipp BR, Voss JS, Daniel S, Aubry MC, Boland JM. Ciliated muconodular papillary tumors of the lung can occur in Western patients and show mutations in BRAF and AKT1. Am J Surg Pathol. 2016;40:1631–1636. doi: 10.1097/PAS.0000000000000707. [DOI] [PubMed] [Google Scholar]
- 6.Jin Y, Shen X, Shen L, Sun Y, Chen H, Li Y. Ciliated muconodular papillary tumor of the lung harboring ALK gene rearrangement: Case report and review of the literature. Pathol Int. 2017;67:171–175. doi: 10.1111/pin.12512. [DOI] [PubMed] [Google Scholar]
- 7.Kogita A, Yoshioka Y, Sakai K, Togashi Y, Sogabe S, Nakai T, Okuno K, Nishio K. Inter- and intra-tumor profiling of multi-regional colon cancer and metastasis. Biochem Biophys Res Commun. 2015;458:52–56. doi: 10.1016/j.bbrc.2015.01.064. [DOI] [PubMed] [Google Scholar]
- 8.Sato S, Koike T, Homma K, Yokoyama A. Ciliated muconodular papillary tumour of the lung: a newly defined low-grade malignant tumour. Interact Cardiovasc Thorac Surg. 2010;11:685–687. doi: 10.1510/icvts.2009.229989. [DOI] [PubMed] [Google Scholar]
- 9.Hata Y, Yuasa R, Sato F, Otsuka H, Goto H, Isobe K, Mitsuda A, Wakayama M, Shibuya K, Takagi K, Watanabe Y. Ciliated muconodular papillary tumor of the lung: a newly defined low-grade malignant tumor with CT findings reminiscent of adenocarcinoma. Jpn J Clin Oncol. 2013;43:205–207. doi: 10.1093/jjco/hys218. [DOI] [PubMed] [Google Scholar]
- 10.Taguchi R, Higuchi K, Sudo M, Kenji M, Miyamoto T, Mishima O, Kitano M, Azuhata K, Ito N. A case of anaplastic lymphoma kinase (ALK)-positive ciliated muconodular papillary tumor (CMPT) of the lung: Case report and review of the literature. Pathol Int. 2017;67:99–104. doi: 10.1111/pin.12504. [DOI] [PubMed] [Google Scholar]
- 11.Kamata T, Yoshida A, Kosuge T, Watanabe S, Asamura H, Tsuta K. Ciliated muconodular papillary tumors of the lung: a clinicopathologic analysis of 10 cases. Am J Surg Pathol. 2015;39:753–760. doi: 10.1097/PAS.0000000000000414. [DOI] [PubMed] [Google Scholar]
- 12.Kon T, Baba Y, Fukai I, Watanabe G, Uchiyama T, Murata T. Ciliated muconodular papillary tumor of the lung: a Report of five cases. Pathol Int. 2016;66:633–639. doi: 10.1111/pin.12460. [DOI] [PubMed] [Google Scholar]
- 13.Chu HH, Park SY, Cha EJ. Ciliated muconodular papillary tumor of the lung: The risk of false-positive diagnosis in frozen section. Hum Pathol Case Reports. 2017;7:8–10. doi: 10.1016/j.ehpc.2015.08.003. [DOI] [Google Scholar]
- 14.Chuang HW, Liao JB, Chang HC, Wang JS, Lin SL, Hsieh PP. Ciliated muconodular papillary tumor of the lung: a newly defined peripheral pulmonary tumor with conspicuous mucin pool mimicking colloid adenocarcinoma: a case report and review of literature. Pathol Int. 2014;64:352–357. doi: 10.1111/pin.12179. [DOI] [PubMed] [Google Scholar]
- 15.Lau KW, Aubry MC, Tan GS, Lim CH, Takano AM. Ciliated muconodular papillary tumor: a solitary peripheral lung nodule in a teenage girl. Hum Pathol. 2016;49:22–26. doi: 10.1016/j.humpath.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa M, Sumitomo S, Imamura N, Nishida T, Mineura K, Ono K. Ciliated muconodular papillary tumor of the lung: report of five cases. J Surg Case Rep. 2016;2016:rjw144. doi: 10.1093/jscr/rjw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaufmann O, Dietel M. Thyroid transcription factor-1 is the superior immunohistochemical marker for pulmonary adenocarcinomas and large cell carcinomas compared to surfactant proteins A and B. Histopathology. 2000;36:8–16. doi: 10.1046/j.1365-2559.2000.00801.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data is available upon request to the corresponding author.


