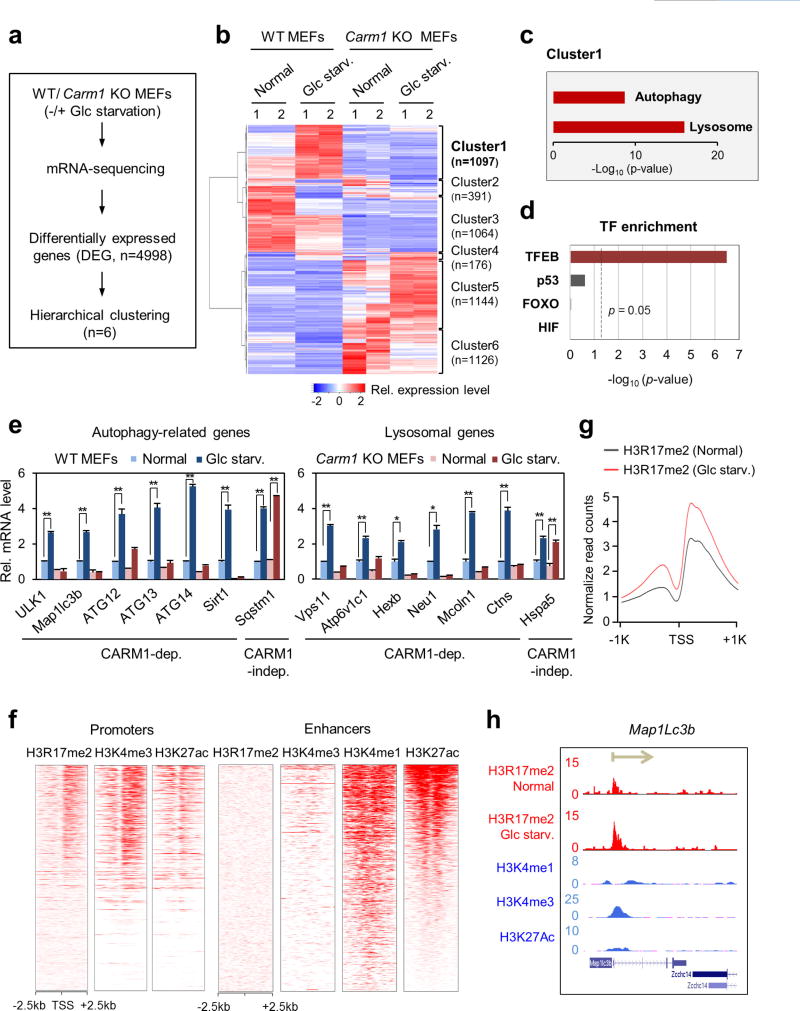
Extended Data Figure 6. Identification of CARM1 target genes by RNA-seq and ChIP–seq analyses.
a, Flow chart showing the strategy of RNA-seq analysis. b, Hierarchical clustering results applied to 4,998 differentially expressed genes (DEGs). c, Autophagy-related and lysosomal genes significantly observed in cluster 1. Hyper-geometric P values were calculated. d, Genes from cluster 1 were analysed for transcription factor (TF) motif enrichment at their promoter region (− 500–100). Hypergeometric P values were calculated. e, qRT–PCR analysis of CARM1-dependent autophagy-related and lysosomal genes. Data are mean ± s.e.m.; n = 3. *P < 0.05, **P < 0.01 (one-tailed t-test). f, Enrichment of H3R17me2 at promoters (left) and enhancers (right). The data on H3R17me2, H3K4me1, H3K4me3 and H3K27ac were obtained from MEFs under normal condition. g, Increase in H3R17me2 at promoters of genes from cluster 1 after glucose starvation. h, Increased H3R17me2 levels in response to 18 h of glucose starvation at the autophagy-related gene Map1lc3b. The direction of transcription is indicated by the arrow and the beginning of the arrow indicates the TSS.