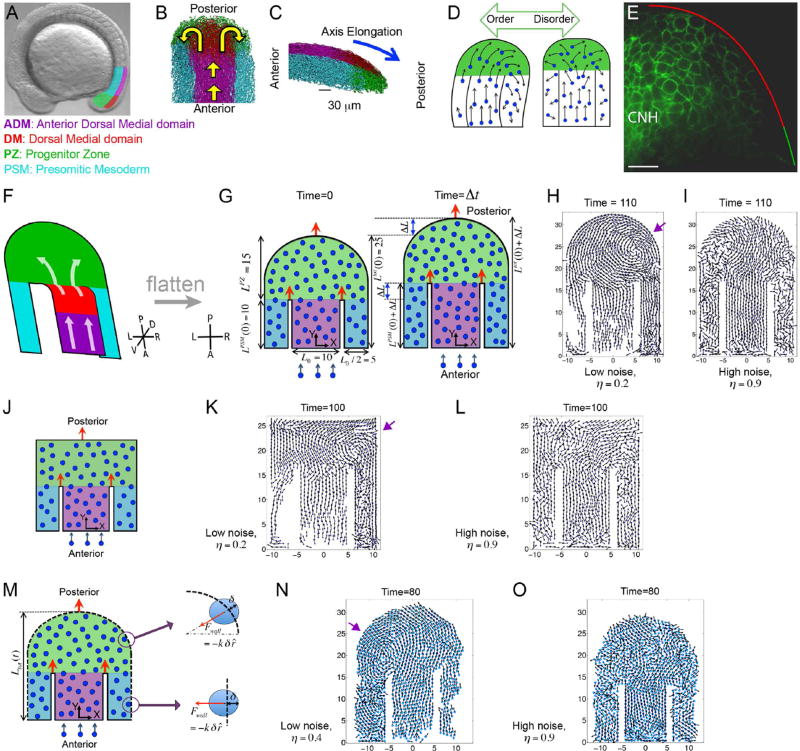
Figure 1. An order to disorder transition in in vivo and in silico tailbuds.
(A) A zebrafish embryo with four tailbud domains highlighted: ADM (magenta), DM (red), PZ (green) and PSM (cyan). (B–C) Dorsal (B) and lateral (C) views of experimental cell tracks within the tailbud. Yellow arrows in B indicate the overall flow direction, while the blue arrow in C denotes the direction of posterior elongation. (D) A schematic posing our hypothesis that overly ordered motion in the PZ breaks axial symmetry. (E) A mid-sagittal view of Cadherin 2-GFP localization along the boundary between the DM and PZ. Anterior is to the left. The chordoneural hinge (CNH) expresses very high levels of Cadherin 2. Posterior to the CNH, Cadherin 2 levels are substantially higher in the DM (red) than in the PZ (green). n= 9 embryos. Scale bar equals 20 microns. (F) The general flow of cells in the tailbud represented as a folded 2D surface. Our computer model is a flattened version of this flow pattern. (G) A diagram of our 2D model involving self-propelled particles within a horseshoe geometry with rigid boundaries. The dimensions reflect the in vivo data, and domains are colored as in panels A-C, but the ADM and DM are merged into a single domain (purple). Left: The tailbud at the initial time t = 0. The widths of the ADM and PSM, and the initial length are shown. Right: The tailbud at a later time Δt. After time Δt, the lengths incrementally increase by an amount ΔL. Orange arrows indicate the posterior movements of the boundaries. New particles are introduced at a fixed rate into the ADM. In simulations (Movie S1), vortices emerge (purple arrow) at low noise (H), and PZ cells sort asymmetrically creating left/right population differences. (I) At high noise cells sort symmetrically. (J) An idealized model of the tailbud with rectangular PZ. (K, L) Snapshots of simulated cell dynamics at low (K) and high noise (L). Note the vortex in K (purple arrow). (M) A variation of our 2D model of tailbud with an elastic outer boundary (dashed lines). (N, O) The elastic forces on a cell penetrating the boundary are shown on the right. Simulation snapshots of the cell dynamics at a low noise (N) and at high noise (O) in the presence of elastic boundaries. Note the vortex in N (purple arrow). The times are in units of 36 in silico iterations. Parameters are specified in Methods. See also Movie S1.