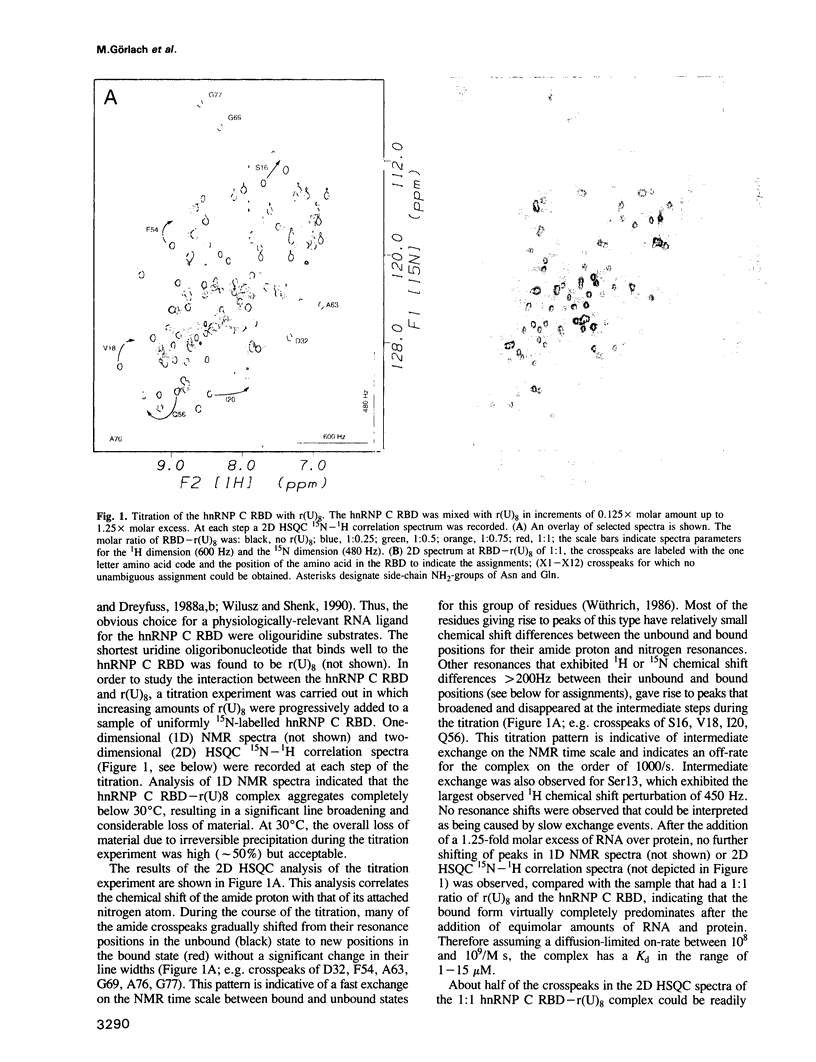
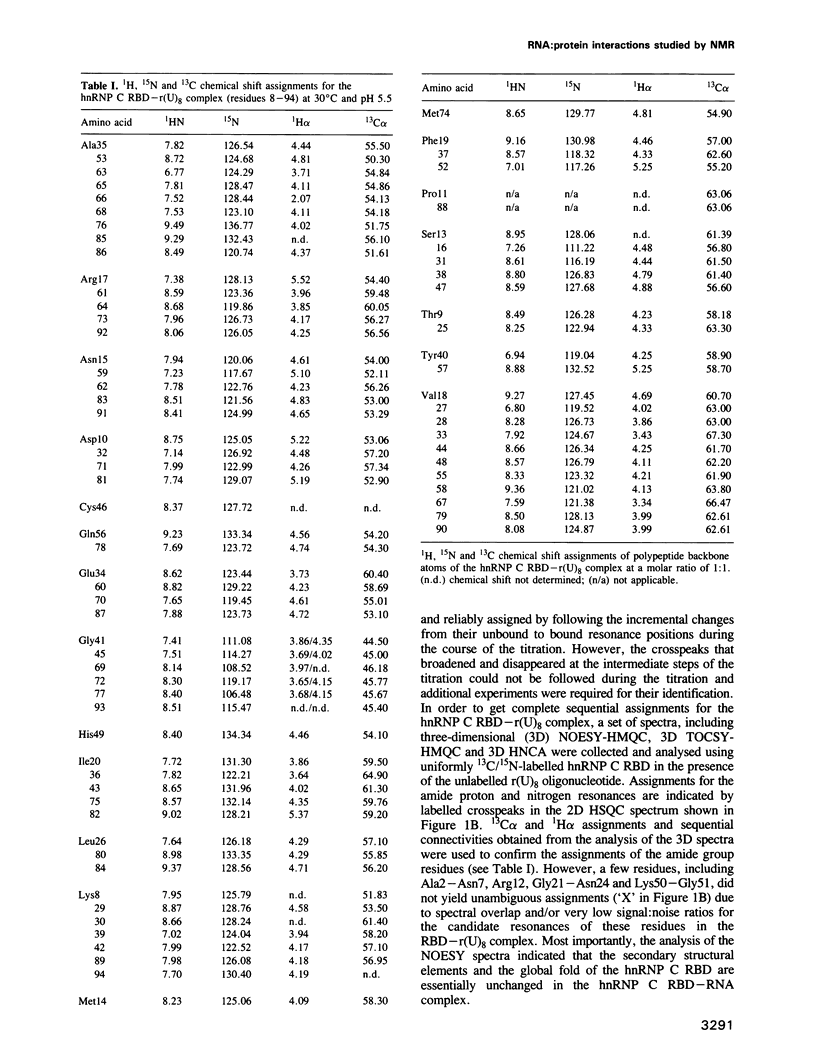
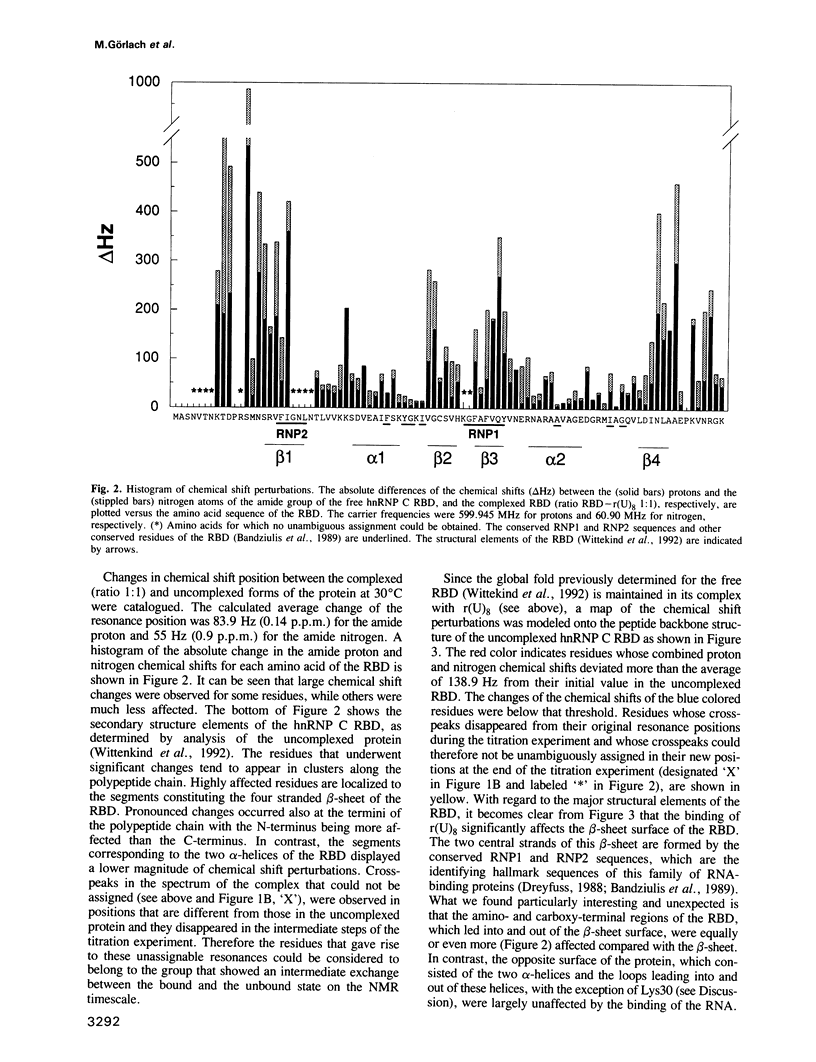
Abstract
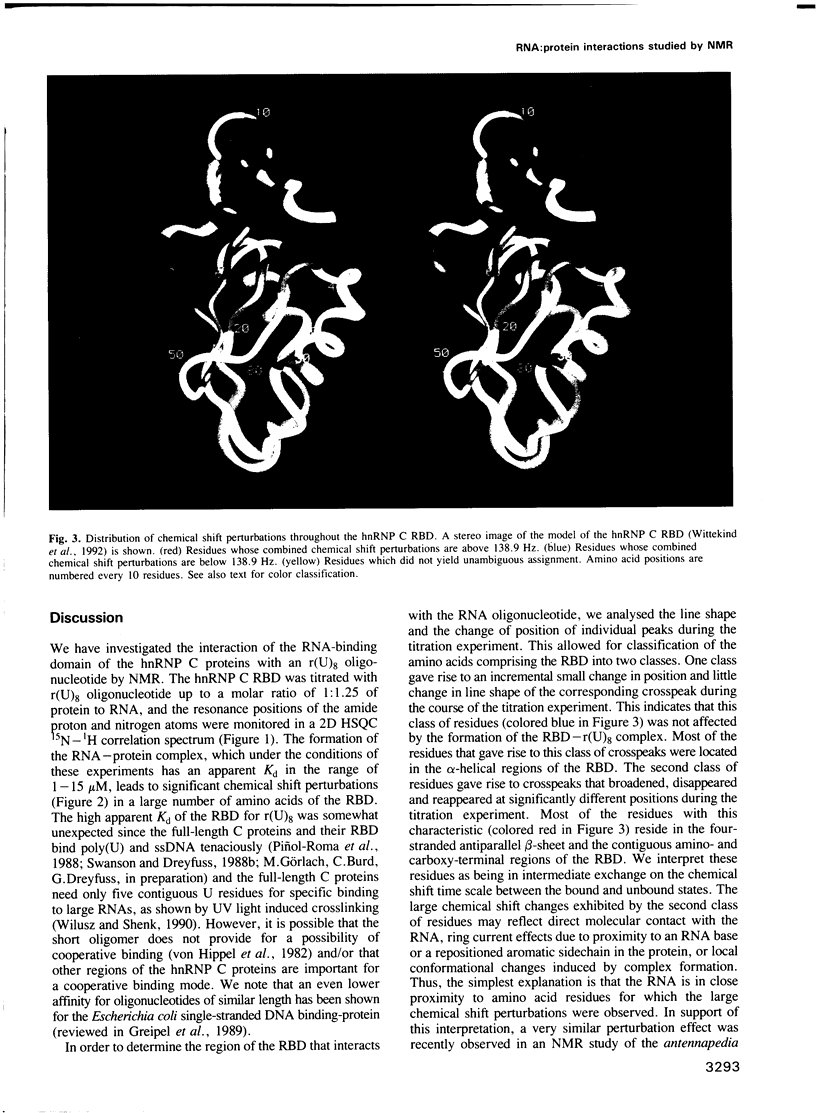
The hnRNP C proteins are among the most abundant and avid pre-mRNA-binding proteins and they contain a consensus sequence RNA-binding domain (RBD) that is found in a large number of RNA-binding proteins. The interaction of the RBD of the hnRNP C proteins with an RNA oligonucleotide [r(U)8] was monitored by nuclear magnetic resonance (NMR). 15N and 13C/15N-labelled hnRNP C protein RBD was mixed with r(U)8 and one- and two-dimensional (1D and 2D) NMR spectra were recorded in a titration experiment. NMR studies of the uncomplexed 93 amino acid hnRNP C RBD (Wittekind et al., 1992) have shown that it has a compact folded structure (beta alpha beta beta alpha beta), which is typical for the RBD of this family of proteins and which is comprised of a four-stranded antiparallel beta-sheet, two alpha-helices and relatively unstructured amino- and carboxy-terminal regions. Sequential assignments of the polypeptide main-chain atoms of the hnRNP C RBD-r(U)8 complex revealed that these typical structural features are maintained in the complex, but significant perturbations of the chemical shifts of amide group atoms occur in a large number of residues. Most of these residues are in the beta-sheet region and especially in the terminal regions of the RBD. In contrast; chemical shifts of the residues of the well conserved alpha-helices, with the exception of Lys30, are not significantly perturbed. These observations localize the candidate residues of the RBD that are involved in the interaction with the RNA.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthony-Cahill S. J., Benfield P. A., Fairman R., Wasserman Z. R., Brenner S. L., Stafford W. F., 3rd, Altenbach C., Hubbell W. L., DeGrado W. F. Molecular characterization of helix-loop-helix peptides. Science. 1992 Feb 21;255(5047):979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989 Apr;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Burd C. G., Matunis E. L., Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991 Jul;11(7):3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Swanson M. S., Görlach M., Dreyfuss G. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9788–9792. doi: 10.1073/pnas.86.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvoli M., Cobianchi F., Biamonti G., Riva S. Recombinant hnRNP protein A1 and its N-terminal domain show preferential affinity for oligodeoxynucleotides homologous to intron/exon acceptor sites. Nucleic Acids Res. 1990 Nov 25;18(22):6595–6600. doi: 10.1093/nar/18.22.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. D., Grabowski P. J., Sharp P. A., Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986 Mar 28;231(4745):1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- Cobianchi F., SenGupta D. N., Zmudzka B. Z., Wilson S. H. Structure of rodent helix-destabilizing protein revealed by cDNA cloning. J Biol Chem. 1986 Mar 15;261(8):3536–3543. [PubMed] [Google Scholar]
- Davis R. L., Cheng P. F., Lassar A. B., Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990 Mar 9;60(5):733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Swanson M. S., Piñol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci. 1988 Mar;13(3):86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Hoffman D. W., Query C. C., Golden B. L., White S. W., Keene J. D. RNA-binding domain of the A protein component of the U1 small nuclear ribonucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2495–2499. doi: 10.1073/pnas.88.6.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen T. H., Oubridge C., Teo C. H., Pritchard C., Nagai K. Identification of molecular contacts between the U1 A small nuclear ribonucleoprotein and U1 RNA. EMBO J. 1991 Nov;10(11):3447–3456. doi: 10.1002/j.1460-2075.1991.tb04909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Kenan D. J., Query C. C., Keene J. D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991 Jun;16(6):214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- Kumar A., Williams K. R., Szer W. Purification and domain structure of core hnRNP proteins A1 and A2 and their relationship to single-stranded DNA-binding proteins. J Biol Chem. 1986 Aug 25;261(24):11266–11273. [PubMed] [Google Scholar]
- Lutz-Freyermuth C., Query C. C., Keene J. D. Quantitative determination that one of two potential RNA-binding domains of the A protein component of the U1 small nuclear ribonucleoprotein complex binds with high affinity to stem-loop II of U1 RNA. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6393–6397. doi: 10.1073/pnas.87.16.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion D., Driscoll P. C., Kay L. E., Wingfield P. T., Bax A., Gronenborn A. M., Clore G. M. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemistry. 1989 Jul 25;28(15):6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W. A binding consensus: RNA-protein interactions in splicing, snRNPs, and sex. Cell. 1989 Apr 7;57(1):1–3. doi: 10.1016/0092-8674(89)90164-5. [DOI] [PubMed] [Google Scholar]
- Merrill B. M., Stone K. L., Cobianchi F., Wilson S. H., Williams K. R. Phenylalanines that are conserved among several RNA-binding proteins form part of a nucleic acid-binding pocket in the A1 heterogeneous nuclear ribonucleoprotein. J Biol Chem. 1988 Mar 5;263(7):3307–3313. [PubMed] [Google Scholar]
- Nagai K., Oubridge C., Jessen T. H., Li J., Evans P. R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990 Dec 6;348(6301):515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- Nietfeld W., Mentzel H., Pieler T. The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J. 1990 Nov;9(11):3699–3705. doi: 10.1002/j.1460-2075.1990.tb07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil K. T., Hoess R. H., DeGrado W. F. Design of DNA-binding peptides based on the leucine zipper motif. Science. 1990 Aug 17;249(4970):774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- Otting G., Qian Y. Q., Billeter M., Müller M., Affolter M., Gehring W. J., Wüthrich K. Protein--DNA contacts in the structure of a homeodomain--DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990 Oct;9(10):3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S., Choi Y. D., Matunis M. J., Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988 Feb;2(2):215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- Query C. C., Bentley R. C., Keene J. D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989 Apr 7;57(1):89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- Scherly D., Boelens W., Dathan N. A., van Venrooij W. J., Mattaj I. W. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B'' and their cognate RNAs. Nature. 1990 Jun 7;345(6275):502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- Scherly D., Boelens W., van Venrooij W. J., Dathan N. A., Hamm J., Mattaj I. W. Identification of the RNA binding segment of human U1 A protein and definition of its binding site on U1 snRNA. EMBO J. 1989 Dec 20;8(13):4163–4170. doi: 10.1002/j.1460-2075.1989.tb08601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988 May;8(5):2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3' end of introns. EMBO J. 1988 Nov;7(11):3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J., Feig D. I., Shenk T. The C proteins of heterogeneous nuclear ribonucleoprotein complexes interact with RNA sequences downstream of polyadenylation cleavage sites. Mol Cell Biol. 1988 Oct;8(10):4477–4483. doi: 10.1128/mcb.8.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J., Shenk T. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol Cell Biol. 1990 Dec;10(12):6397–6407. doi: 10.1128/mcb.10.12.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind M., Reizer J., Deutscher J., Saier M. H., Klevit R. E. Common structural changes accompany the functional inactivation of HPr by seryl phosphorylation or by serine to aspartate substitution. Biochemistry. 1989 Dec 26;28(26):9908–9912. doi: 10.1021/bi00452a005. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Zuiderweg E. R., Fesik S. W. Heteronuclear three-dimensional NMR spectroscopy of the inflammatory protein C5a. Biochemistry. 1989 Mar 21;28(6):2387–2391. doi: 10.1021/bi00432a008. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Kowalczykowski S. C., Lonberg N., Newport J. W., Paul L. S., Stormo G. D., Gold L. Autoregulation of gene expression. Quantitative evaluation of the expression and function of the bacteriophage T4 gene 32 (single-stranded DNA binding) protein system. J Mol Biol. 1982 Dec 25;162(4):795–818. doi: 10.1016/0022-2836(82)90548-4. [DOI] [PubMed] [Google Scholar]