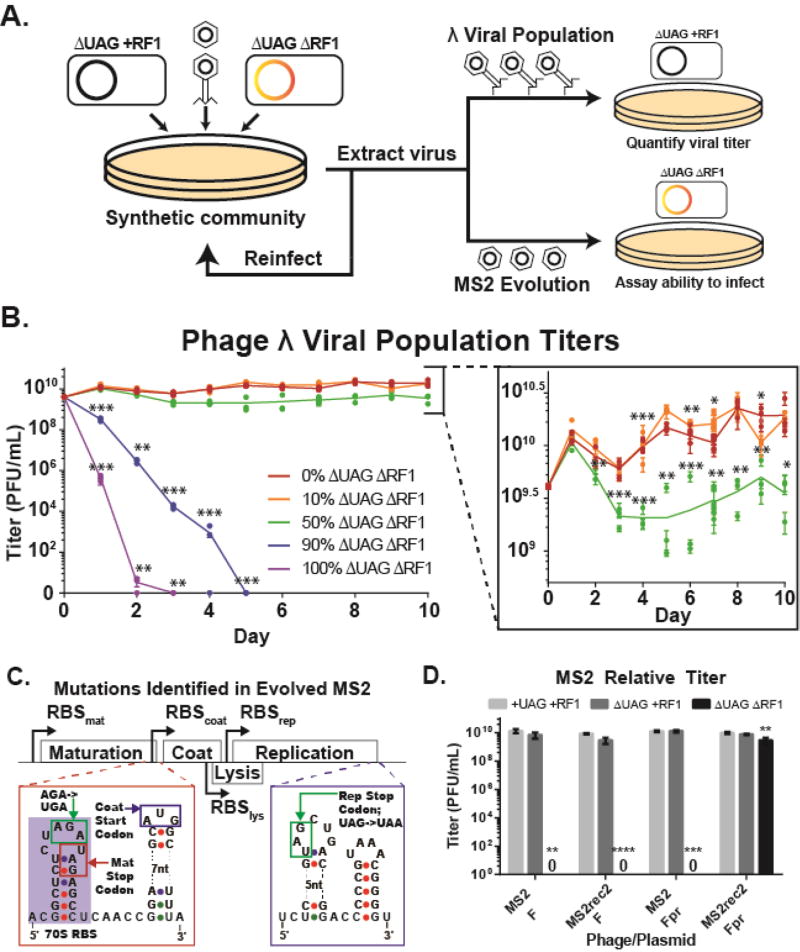
Figure 3. Recoded organisms reduce viral population fitness in microbial communities and select for viral mutations that eliminate UAG codon use.
(A) Schematic of microbial community assays. Phages are infected on a co-culture containing varying ratios of ΔUAG+RF1 and ΔUAGΔRF1, extracted the next day, and propagated on a co-culture with the same cell ratio. Viral populations of λ were quantified by infection on ΔUAG+RF1, and ability of phage MS2 to infect ΔUAGΔRF1 was assayed by plating on ΔUAGΔRF1 containing pFpr. (B) Titers of phage λ viral populations propagated on microbial communities containing cells with standard and alternate genetic codes. Lines are mean of 3 biological replicates for each population. (C) Location of mutations eliminating UAG codon usage in the MS2 genome (Calendar, 2006; Fiers et al., 1976). (D) Relative titers of wild-type and recoded MS2 (MS2rec2) phages infected on ΔUAGΔRF1 with pF or pFpr, which is required for phage infection. Data are mean with standard deviation, n=3. P-values are as follows: * is P ≤ 0.05, ** is P ≤ 0.01, *** is P ≤ 0.001, and **** is P ≤ 0.0001.