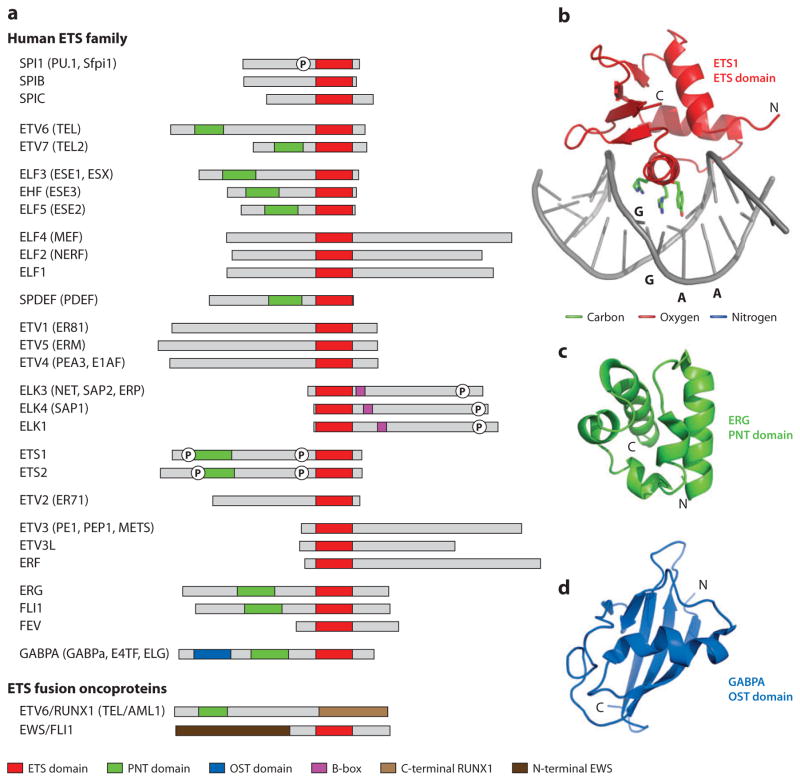
Figure 1.
Structural and functional domains of the ETS family of transcription factors. (a) Nomenclature and domain organization of the 28 paralogous human ETS proteins (grouped according to the dendogram of Figure 2). We use HUGO nomenclature (191) for all ETS genes and proteins, with alternative names provided. Oncogenic ETS proteins, which are created by human chromosome rearrangements, are illustrated by two examples. Many ETS genes produce multiple protein products via alternative splicing/start sites, and in these cases, a single polypeptide was chosen arbitrarily. Boxes identify the DNA-binding ETS domain (red), PNT domain (green), OST domain (blue), and B-box (magenta) of the ETS factors as well as the C-terminal portion of RUNX1 (light brown) and the N-terminal portion of EWS (dark brown). The circled P symbolizes a phosphorylated region discussed in the text. Not identified are additional regions involved in transcriptional activation/repression, posttranslational modifications (including kinase docking and sumoylation), nuclear import/export, turnover, and so forth. (b) Ribbon diagram of the ETS1 ETS domain bound to a DNA duplex (1k79.pdb with the inhibitory helices deleted for clarity). The side chains of R391, R394, and Y395, which provide base-specific contacts to core GGAA, are also shown in stick format. Ribbon diagrams of (c) the ERG PNT domain (1sxe.pdb) and (d) the GABPA OST domain (1juo.pdb). See Supplemental Tables 2 and 3 for a referenced summary of all published ETS and PNT domain structures. Molecular diagrams were rendered with PyMOL (http://pymol.org) with coordinate files from the Protein Data Bank (http://pdb.org).