Figure 1. Assembly of CRL3GCL is required for proper PGC formation in Drosophila.
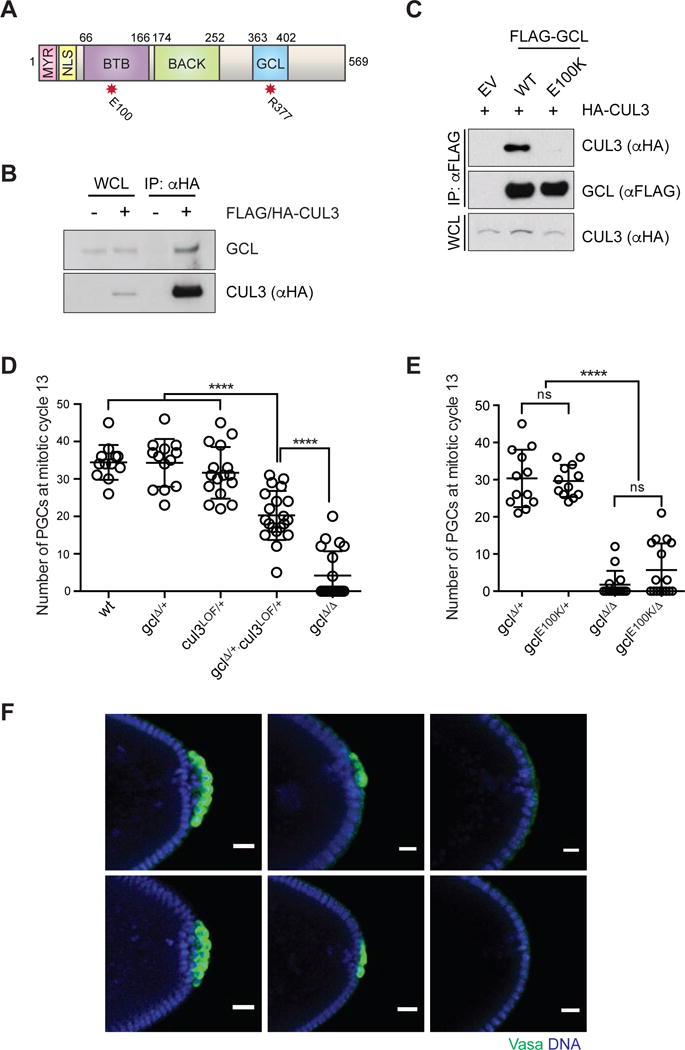
(A) Domain architecture of GCL protein. MYR: myristoylation signal, NLS: nuclear localization signal, BTB: Broad-Complex, Tramtrack and Bric a brac domain, BACK: BTB and C-terminal Kelch domain) and the conserved GCL domain. The red asterisks highlight the position of functional mutants used in this study.
(B) Ovary lysates were prepared from females expressing FLAG-HA-tagged CUL3 (driven by UAS promoter using the germline-specific driver nanos-Gal4) and immunoprecipiated with anti-HA resin. Immuno-complexes were probed with an antibody recognizing endogenous GCL. IP, immunoprecipitation; WCL, whole cell lysate.
(C) Drosophila Schneider 2 (S2) cells were co-transfected with the indicated constructs or empty vector (EV), as indicated. Twenty-four hours after transfection, cells were harvested and lysed. cell lysates were immunoprecipitated (IP) with anti-FLAG resin, and immunocomplexes were probed with indicated antibodies.
(D, E) Number of PGCs in embryos of indicated maternal genotype. Each circle represents the number of PGCs counted in a single embryo. Bars represent the mean ± standard deviation. (n > 12, ****P < 0.0001, ns = not significant, Mann-Whitney test)
(F) Immunostaining of embryos from females of indicated genotype for expression of PGC marker Vasa (green). DAPI for DNA (blue). Posterior poles of representative embryos are shown. Scale bar = 20μm.
See also Figure S1.
