Summary
Oculocutaneous albinism Type 1 (OCA1) is an autosomal recessive disorder caused by mutations in the tyrosinase gene. Two subtypes of OCA1 have been described: severe OCA1A with complete absence of tyrosinase activity and less severe OCA1B with residual tyrosinase activity. Here, we characterize the recombinant human tyrosinase intra-melanosomal domain and mutant variants, which mimic genetic changes in both subtypes of OCA1 patients. Proteins were prepared using site-directed mutagenesis, expressed in insect larvae, purified by chromatography, and characterized by enzymatic activities- tryptophan fluorescence, and Gibbs free energy changes. The OCA1A mutants show very low protein expression, protein yield, and are enzymatically inactive. Mutants mimicking OCA1B were biochemically similar to the wild type, but exhibited lower specific activities and protein stabilities. The results are consistent with clinical data, which indicates that OCA1A mutations inactivate tyrosinase and result in severe phenotype, while OCA1B mutations partially inactive tyrosinase and results in OCA1B albinism.
Keywords: oculocutaneous albinism, tyrosinase, genetic mutations, protein structure, protein stability, protein unfolding, protein purification
Introduction
Albinism is a heterogeneous group of genetic disorders currently defined by abnormalities in visual system development and a variable hypopigmentation phenotype (Montoliu et al., 2014). There are two major types of albinism: non-syndromic and syndromic. Non-syndromic albinism includes X-linked ocular albinism (OA), caused by mutation in GPR143 (OA1) gene that is confined to the eyes, and oculocutanious albinism (OCA), in which lack or reduction of pigment affects the eyes, skin and hair (Schiaffino and Tacchetti, 2005, Gronskov et al., 2007). There are several subtypes of OCA caused by genetic mutations in the following genes: TYR (Oculocutaneous albinism type 1, OCA1), OCA2 (P gene, OCA2), TYRP1 (OCA3), SLC45A2 (OCA4), OCA5 (OCA5), SLC24A5 (OCA6), C10ORF11 (OCA7) (Tripathi et al., 1992b, Manga et al., 1997, King et al., 1978, King et al., 1986, Boissy et al., 1996, Newton et al., 2001, Gronskov et al., 2013, Simeonov et al., 2013, Wei et al., 2013). Syndromic forms of albinism, such as Hermansky-Pudlak (types 1–10) and the Chediak-Higashi syndromes, are caused by mutations in HPS1–10 and LYST genes, respectively. They are characterized by similar clinical phenotypes as OCA, but have additional health perturbations (Oh et al., 1998, Jung et al., 2006, Anikster et al., 2001, Suzuki et al., 2002, Zhang et al., 2003, Huizing et al., 2009, Li et al., 2003, Morgan et al., 2006, Cullinane et al., 2011, Ammann et al., 2016). These syndromes have different levels of severity, commonness and occurrence in ethnic groups (OMIM®, An Online Catalog of Human Genes and Genetic Disorders; http://www.omim.org).
OCA1, the most common type of albinism, is caused by bi-allelic mutations (missense, nonsense or frameshift) in the TYR gene on chromosome 11q14.3 and occurs in approximately 1: 40,000 worldwide (total OCA – 1:17,000). This type of albinism, most common in Caucasians, is subdivided clinically into two groups: the most severe type of albinism, in which tyrosinase activity and melanin synthesis are undetectable, OCA1A (OMIM # 203100), or partially measurable, OCA1B (OMIM # 606952). Clinically, OCA1A patients have a life-long absence of melanin, and presentwhite hair and eyelashes, milky skin, and light blue or translucent-appearing (red in bright light) irides. In OCA1B patient’s symptoms are present perinatally, but may improve with age. Both subtypes of OCA1 result in reduced best-corrected visual acuity, nystagmus, foveal hypoplasia, and abnormal decussation of ganglion cell axons at the optic chiasm (Oetting, 2000). OCA1 was suggested to be an endoplasmic reticulum (ER) retention disease in which misfolded tyrosinase mutants are retained in the ER by cellular quality control (Halaban et al., 2001, Popescu et al., 2005).
Nearly 350 mutations were found dispersed throughout the TYR gene, which involves approximately 77% missense mutations (OCA1A – 67% and OCA1B – 33% from patients with established diagnosis), ~15% deletions, and ~3% insertions (HGMD Professional 2016.2 database, https://portal.biobase-international.com/hgmd/pro/gene.php?gene=TYR). These mutations affect the normal production of melanin either by full or partial elimination of the tyrosinase activity.
Tyrosinase is a membrane glycoprotein widely expressed in plant/animal tissues and crucial in melanin production (Wang and Hebert, 2006, Bijelic et al., 2015). The enzyme catalyzes the oxidation of L-tyrosine (monophenol substrate) and L- 3, 4-dihydroxyphenylalanine (L-DOPA, diphenol substrate) to form dopaquinone (Riley, 1999). Although typical Michaelis-Menten behavior is observed for both monophenol and diphenol activities, the Km values for each substrate ranges widely across species, e.g. for L-DOPA, Km values 0.016 mM (Mus musculus, (Hearing et al., 1978)) to 8.1 mM (Streptomyces, (Kohashi et al., 2004)). The monophenolase and diphenol oxidase activities are linked to the tyrosinase active site, which is composed of six histidine residues that coordinate two copper ions (CuA and CuB) essential for activity (Oetting, 2000, King et al., 2003). Previous data show that activity and stability of mutant tyrosinase could be indirectly improved with using nitisinone, an FDA –approved inhibitor of tyrosine degradation, by increasing the concentration of substrate (tyrosine) available to the enzyme (Onojafe et al., 2011). Human tyrosinase (OMIM*606933) is post-translationally modified in the ER by addition of several N-linked glycans, which are required for protein maturation and stability (Imokawa and Mishima, 1984). Glycosylation appears to have a crucial role in the translocation of protein from ER to the cytoplasm (Wang and Hebert, 2006). Human tyrosinase is predicted to have 28 asparagine residues (N), of which 7 (N-86, -111, -161, -230, -290, -337, and -371) have the N-x-S/T sequence motif for potential N-glycosylation.
We have previously shown that a recombinant intra-melanosomal domain of human tyrosinase (hTyrCtr) and two temperature-sensitive OCA1B-related mutant variants are soluble monomeric glycoproteins with enzymatic activities mirroring the in vivo function (Dolinska et al., 2014). Also, it was shown that hTyrCtr contains at least five N-glycosylation sites, which are either completely (N86, N337) or partially (N111, N230, N290) modified ((Dolinska et al., 2014), see also Figure 1).
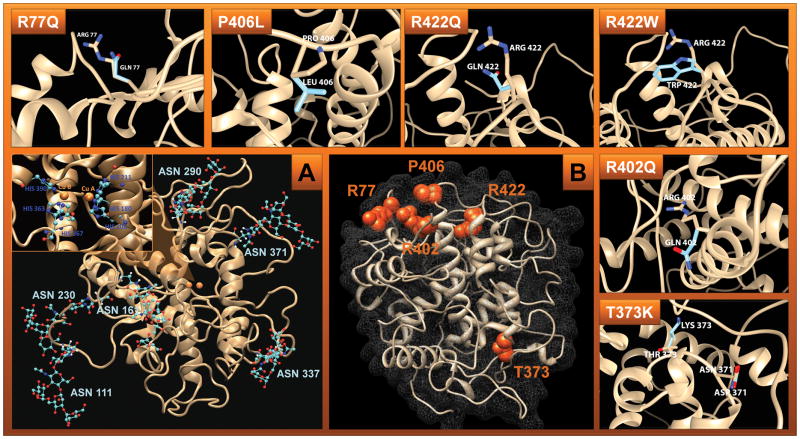
Figure 1. Protein structure of N-glycosylated wild-type Tyr.
Panel A: Glycosylated Tyr. The protein backbone is depicted as a beige cartoon ribbon. Glycans are shown attached to their respective asparagine side chains in a ball-and-stick model with red indicating oxygen, cyan indicating carbon, and blue indicating nitrogen. The Insert shows the active site. The two copper atoms (copper A and copper B) are depicted as orange spheres in the center of the active site, bridged by a hydroxide ion. Cu A is coordinated by the epsilon nitrogen of HIS 180, HIS 202, and HIS 211. Cu B is coordinated by the epsilon nitrogen of HIS 363, HIS 367, and HIS 390.
Panel B: Residues undergo OCA1-related mutations (orange spheres representing their side chain atoms) mapped to the human tyrosinase protein structure model as described in methods section. The protein backbone structure is depicted as a beige cartoon ribbon and the protein surface is shown by grey mesh. Surrounding rectangles show Tyr mutants superimposed over wild type. R77Q, R402Q, and R422Q – the glutamine mutation is superimposed over the wild type amino acid residue, arginine. R422W - the tryptophan mutation is superimposed over the wild type amino acid residue, arginine. P406L – the leucine mutation is superimposed over the wild type amino acid residue, proline. The wild-type residues are depicted in beige and the mutants are depicted in baby blue. The protein backbones are shown as a beige-colored cartoon ribbon.
In this work, we propose that the genetic mutations in tyrosinase, which give rise to OCA1 albinism, mainly influence protein folding, stability, and intrinsic activity. This link between structural change and severity of mutation was studied at the protein level by biochemically and biophysically characterizing a number of mutant variants of hTyrCtr that mimic OCA1 genetic changes. All mutant variants were selected based on available patient information from the National Institutes of Health. These include: OCA1A-related mutants: T373K and R77Q, and OCA1B-related mutations, R402Q, R422W, R422Q, and P406L. The OCA1A mutants were unstable and showed no enzymatic activity. Mutants mimicking OCA1B were biochemically similar to the wild type, however, showed decreased enzymatic activity and protein stability.
Results
Expression and purification of hTyrCtr and OCA1-related mutants
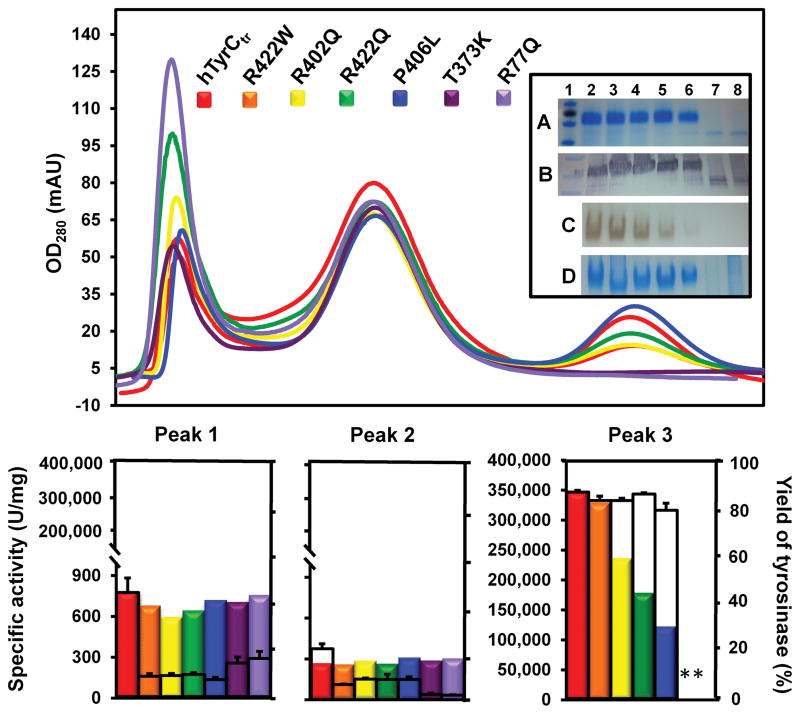
Protein expression of hTyrCtr and its OCA1-related mutants was assessed by SDS-PAGE and Western blot of protein supernatants from larval lysates; all showed fairly similar protein levels (Supplementary Materials, Figure S1). At the second purification stage (Sephacryl S-300 HR), three peaks were observed: Peak 1, which corresponds to the void volume of the column (exclusion limit ~2000 kDa), Peak 2, proteins in the 20 – 300 kDa polypeptide range with a very low tyrosinase activity, and Peak 3, which matches elution of pure hTyrCtr and OCA1B mutants (Figure 2). The SDS-PAGE and Western blots of the chromatography peaks shown in Figure 2 (insert lanes A, B) and native gel electrophoresis (insert lanes C, D) shows that hTyrCtr and OCA1B-related mutants migrated well into the gel (not aggregated) and reacted strongly with L-DOPA. Based on these results, the hTyrCtr and OCA1B-related mutants were purified as soluble proteins with high enzymatic activities.
Figure 2. Purification and enzymic activity of recombinant hTyrCtr and OCA1-related mutant variants.
Top panel: Absorbance values recorded at 280 nm after two steps of purification using ÄKTAxpress chromatography system. Chromatography of hTyrCtr (red) and mutant variants (R422W, orange; R402Q, yellow; R422Q, green; P406L, blue; T373K, purple, and R77Q, light violet) were eluted from Sephacryl S-300 HR column as three peaks, Peak 1 (fractions A2–A9), Peak 2 (B8–C9), and Peak 3 (D8–F9). Insert shows the SDS-PAGE (A), Western blot (B), native gel incubated with 3 mM L-DOPA (C), and native gel stained with GelCode Blue reagent (D) of hTyrCtr, and OCA1-related mutants. From the left: 1, protein ladder; 2, hTyrCtr; 3, R422W; 4, R402Q; 5, R422Q; 6, P406L; 7, T373K; 8, R77Q.
Bottom panel: Specific activity of tyrosinase in the three peaks obtained as the L-DOPA enzyme activity multiplied by the sample total volume, and divided by total protein are shown as the colored bars. The tyrosinase content (yield) of hTyrCtr and mutant variants in all three peaks calculated from the SDS-PAGE gels using UN-SCAN-IT gel ™ gel analysis software are showed as the empty bars.
* Indicate no bands at the proper position on SDS-PAGE gel or no activity.
The hTyrCtr protein and OCA1B-related mutants, R422W, R402Q, R422Q, and P406L. were then applied to a Superose 12 10/300 GL column and eluted as single monomeric peaks (Supplementary Materials, Figure S2A). Previously, sedimentation equilibrium was used to show that hTyrCtr was monomeric with a molecular weight of 56.63 kDa (Dolinska et al., 2014).
The proteins were all >97% pure according to the SDS-PAGE (Table 1). However, the proteins did exhibit broad, heterogeneous bands between 55 and 70 kDa due to glycosylation (Figure 2, insert, panel A). These bands all reacted strongly with the T311 human tyrosinase antibody on the Western blot (Figure 2, insert, panel B).
Table 1.
Enzymatic properties of recombinant hTyrCtr and OCA1B-related mutant variants.
| Protein | Final purity (%) | Specific activity (U/mg) | |||
|---|---|---|---|---|---|
| native | % of hTyrCtr | refolded | % of native | ||
| hTyrCtr | 97.22 ± 1.83 | 347,370 ± 13,497 | - | 332,553 ± 16,102 | 95.73 |
| R422W | 98.56 ± 1.28 | 329,499 ± 41,662 | 94.86 | 299,423 ± 6,800 | 90.87 |
| R402Q | 97.54 ± 0.73 | 236,668 ± 17,367 | 68.13 | 206,060 ± 10,713 | 87.07 |
| R422Q | 97.38 ± 1.20 | 178,611 ± 25,991 | 51.42 | 109,849 ± 7,500 | 61.50 |
| P406L | 97.78 ± 0.86 | 122,355 ± 8,072 | 35.22 | 57,728 ± 1,928 | 47.18 |
Protein purity for hTyrCtr and OCA1B-related mutant variants were measured from SDS-PAGE gels as 100% x tyrosinase band intensity/total protein band using UN-SCAN-IT gel ™ gel analysis software (Silk Scientific, Inc., UT). Specific activity was obtained as the diphenol oxidase activity (as described in Materials and Methods section), multiplied by the sample total volume and divided by total protein.
In contrast, OCA1A mutant variants, T373K and R77Q, were mostly in the first, higher-molecular weight peak during gel filtration, with none present in Peak 3; these corresponded to enzymatically-inactive degraded polypeptides (Figure 2, insert lanes 7–8). Moreover, after concentration from Peak 1, the OCA1A mutant proteins were pelleted by low speed centrifugation, indicating that they were highly aggregated and, therefore, excluded from the further analysis.
Enzymatic activity
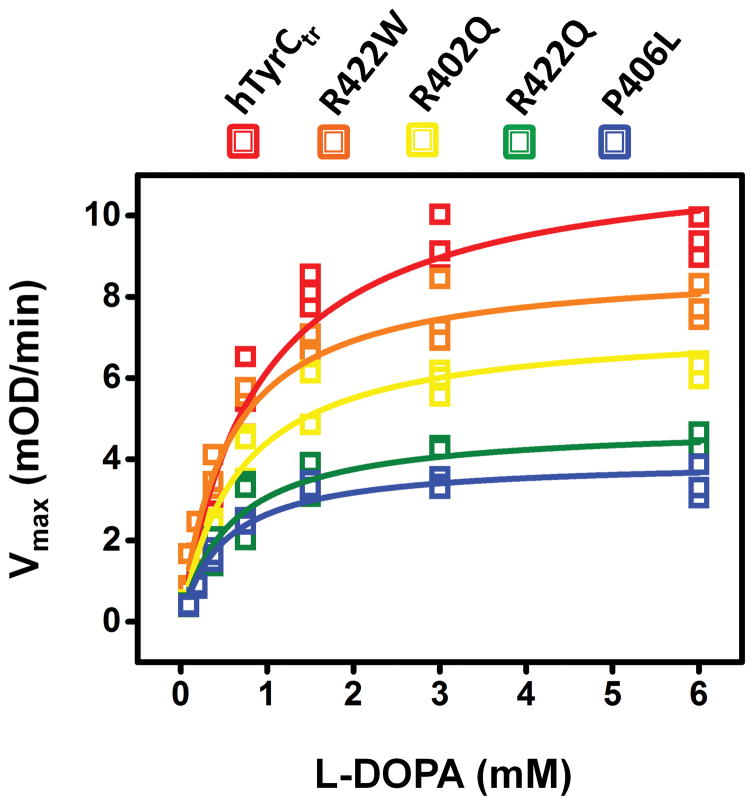
The specific activities of OCA1B-related mutants, R422W, R402Q, R422Q, and P406L, were 95, 68, 51, and 35 % of the hTyrCtr specific activity, respectively (Table 1). Michaelis-Menten constants (Km), which show the concentration of L-DOPA resulting in half of the maximal velocity for the diphenol oxidase reaction, indicated that the binding affinity is slightly higher for the most severe OCA1B-related mutants, R422Q and P406L (Table 2, Figure 3). From the Vmax and Km values, the enzyme turnover, kcat, and enzyme efficiency, kcat/Km, were calculated and indicated that hTyrCtr has higher L-DOPA specificity (150.64) than the OCA1B-related mutants (121.82, 116.07, 107.40, and 111.55 for R422W, R402Q, R422Q, and P406L, respectively) (Table 2). Km values of hTyrCtr and OCA1B mutant variants were similar, indicating similar L-DOPA binding affinities; enzyme turnover was decreased for mutant variants, however.
Table 2.
Kinetics parameters and stability of recombinant hTyrCtr and OCA1B-related mutant variants.
| Protein | Km (mM) | Vmax (mM/sec) | kcat (sec−1) | Kcat/Km (min−1 sec−1) | ΔG (kcal/mol) | ΔΔG |
|---|---|---|---|---|---|---|
| hTyrCtr | 0.85 ± 0.22 | 0.23 ± 0.01 | 128.59 ± 6.52 | 150.64 ± 7.64 | 1.94 ± 0.02 | - |
| R422W | 0.84 ± 0.07 | 0.18 ± 0.03 | 102.57 ± 15.14 | 121.82 ± 17.98 | 1.93 ± 0.07 | −0.01 |
| R402Q | 0.80 ± 0.05 | 0.16 ± 0.01 | 92.63 ± 2.37 | 116.07 ± 2.97 | 2.29 ± 0.05 | +0.35 |
| R422Q | 0.73 ± 0.12 | 0.14 ± 0.02 | 78.44 ± 9.69 | 107.40 ± 13.26 | 2.42 ± 0.07 | +0.48 |
| P406L | 0.65 ± 0.17 | 0.13 ± 0.02 | 72.17 ± 11.07 | 111.55 ± 17.10 | 2.76 ± 0.06 | +0.82 |
Kinetics parameters, the Michaelis-Menten constant (Km) and maximal velocity (Vmax) were obtained from SoftMax Pro software (Molecular Devices, CA) and Vmax was then converted from mOD/min to mM/sec using the rates of kinetics reactions obtained from the initial slopes. The enzyme turnover (kcat) was defined as Vmax/Et, where Et is concentration of enzyme in mM. kcat/Km, enzyme efficiency. ΔG, the Gibbs free energy, was calculated from unfolding/refolding reactions for each protein.
Figure 3. Kinetic analysis of OCA1B-related mutants.
Michaelis-Menten plots of diphenol oxidase activity of hTyrCtr (red), R422W (orange), R402Q (yellow), R422Q (green), and P406L (blue) as a function of L-DOPA concentration measured at 37°C. The lines represent nonlinear fits to the Michaelis-Menten equation obtained from OriginPro software. Experiments were performed in triplicate, and error bars represent the standard deviations.
Conformational and activity changes on urea induced folding - refolding
To test if missense mutations resulted in conformational perturbations, proteins were probed using far-UV circular dichroism, CD (secondary structure) and intrinsic protein fluorescence (tertiary structure). The similarity of the far-UV CD spectra of OCA1B-related mutants and hTyrCtr suggests little or no change in their overall secondary structure (Supplementary Materials, Figure S2B).
The three amino acids give rise to intrinsic fluorescence, phenylalanine (Phe), tyrosine (Tyr), and tryptophan (Trp), but most is due to Trp. The 40 ns tyrosinase homology model indicates that 6 out of a total of 11 Trp are buried inside the enzyme (Supplementary Materials, Figure S3). Thus, Trp can be used to monitor changes in the tertiary structures under unfolding/refolding conditions. Native hTyrCtr has a fluorescence maximum at 340 nm, which upon unfolding with urea red shifts to 363 nm with an increase in intensity (Supplementary Materials, Figures S4 and S5). These spectral changes are consistent with protein unfolding where buried and quenched Trp(s) are solvent exposed. Similar red shifts (23–26 nm) were observed for OCA1B-related mutants after urea treatment (Supplementary Materials, Figure S5). When the denatured protein was refolded, the fluorescence maximum of hTyrCtr was blue-shifted to 340 nm indicating folding reversibility. The OCA1B-related mutants, R422W, R402Q, R422Q, and P406L, when refolded, also blue-shifted to varying degrees, but none reached native-like 340 nm (Table 1, Supplementary Materials, Figures S4 and S5). Although the emission intensity of R422Q was decreased, possibly due to protein aggregation, both mutant variants, R422Q and P406L, showed partially recovered L-DOPA activity after the refolding, 62% and 47%, respectively. Consistent with the fluorescence monitoring, the specific activities of hTyrCtr and mutants R422W and R402Q, exhibited loss and regain of activity, which confirmed the fully reversible denaturation-renaturation cycle. These results are summarized in Table 1.
Equilibrium unfolding and refolding
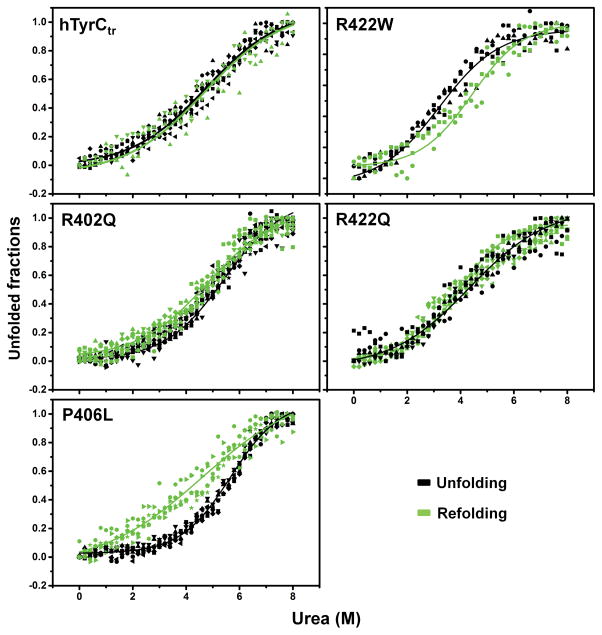
Urea-induced equilibrium unfolding/refolding of hTyrCtr and mutants were obtained by plotting the fluorescence emission 360/320 nm ratio as a function of urea concentration. The sigmoidal curves were fitted using the Boltzmann function (Figure 4). The refolded hTyrCtr, R402Q, and R422Q exhibited native-like response curves whereas the mutant variants, R422W and P406L, curves exhibited hysteresis. The stability of the proteins expressed as free energy changes (ΔG) were determined at zero urea concentration as shown in a Table 2. These changes in protein stability show a remarkable correlation with the specific activity (Figure 5A), suggesting the decrease in protein specific activities in the OCA1B-related missense mutations is due to the decrease in protein stabilities.
Figure 4. Urea-induced equilibrium unfolding/refolding of hTyrCtr and OCA1B-related mutants.
Intrinsic tryptophan fluorescence was measured using an excitation wavelength of 285 nm. Emission spectra were recorded in the range between 300 and 400 nm. Proteins (10 μM) were in 10 mM phosphate buffer, pH 7.4 with 0–8 M urea. For refolding, proteins were dialyzed against 10 mM phosphate buffer, pH 7.4 at 4°C for 24 h. Unfolding and refolding curves were measured as a 360/320 nm ratio and indicated by black and green points, respectively. To visualize the unfolding/refolding the experimental points were fitted with a Boltzmann function using OriginPro 2015 as indicated by black and green lines for unfolding and refolding, respectively.
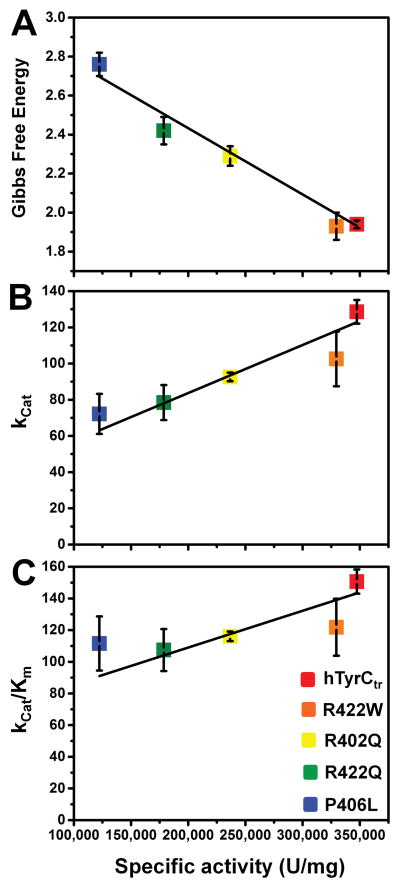
Figure 5. Protein stability, enzyme turnover, and enzyme efficiency as function of specific activity for hTyrCtr and OCA1B-related mutants.
Specific activity of the hTyrCtr and R422W, R402Q, R422Q, and P406L mutant variants was obtained as the diphenol oxidase enzyme activity (as described in Materials and Methods section), multiplied by the sample total volume and divided by total protein.
Panel A shows a negative correlation (Pearson’s r = −0.992) between the experimental Gibbs free energies (ΔG) and the specific activity for each protein. Protein stability ΔG (kcal/mol) values were characterized and calculated from unfolding/refolding reactions.
Panel B represents a positive correlation (Pearson’s r = 0.962) between Kcat, the enzyme turnover (sec−1) and the specific activity. Enzyme turnover was defined as a ratio of Vmax/Et, where Et is concentration of enzyme in mM.
Panel C shows a positive correlation (Pearson’s r = 0.900) between enzyme efficiency, Kcat/Km (min−1 sec−1), and the specific activity.
Catalytic properties of the recombinant human full length and truncated tyrosinases
In order to compare the enzymatic properties of hTyrCtr (Dolinska et al., 2014) and full length detergent associated hTyr tyrosinase, kinetic parameters obtained from reactions with L-DOPA were measured under the same conditions in the presence of 0.1% Triton X-100. The kinetic parameters Km, Vmax, and kcat for hTyr were 0.68 ± 0.03 mM, 0.21 ± 0.04 mM/sec, and 140.04 ± 28.89 sec−1, respectively, and for hTyrCtr, were 0.64 ± 0.03 mM, 0.18 ± 0.03 mM/sec, and 103.23 ± 16.26 sec−1, respectively. Specific activity for hTyr was 238,741 ± 6370 U/mg and for hTyrCtr 439,851 ± 18,736 U/mg.
Discussion
Recombinant human tyrosinase was truncated at the C-terminus to remove the membrane associated region. The truncated protein, hTyrCtr correspond to residues 19 – 469 of the native tyrosinase. The hTyrCtr and OCA1-related mutant variants, R422W, R402Q, R422Q, P406L, T373K, and R77Q were expressed in larval biomass and purified by IMAC and size-exclusion chromatography. Both, OCA1A and OCA1B mutants showed similar expression as the wild type protein, while mutants mimicking OCA1A, T373K and R77Q, were not detectable after two steps of purification and exhibited no enzymatic activity in the peak corresponding to tyrosinase. Initial evaluation of human recombinant tyrosinase from the crude lysate was difficult due to the presence of larval endogenous tyrosinase in the sample and was estimated as 200 mg of total tyrosinase. Finally, purified proteins yields were, in average, 3 mg or less of the recombinant tyrosinase from 20 g of frozen larvae.
The OCA1A causing mutations destabilized the tyrosinase structure, resulting in loss of protein activity. Mutants mimicking OCA1B also resulted in structural destabilization, but to a lesser degree with lower specific activities and protein stabilities compared to wild type. Our work quantifies this link, indicating a negative correlation between specific activity and protein stability and positive correlation between specific activity and enzyme turnover and efficiency (Figure 5, A–C). These findings are consistent with clinical data, which suggests mutations involved in OCA1B albinism reduce tyrosinase activity (OMIM®, An Online Catalog of Human Genes and Genetic Disorders; http://www.omim.org).
OCA1A-related mutants are well known known, especially the T373K variant (c.1118C>A, exon 3), which is the most commonly observed and occurs in ~30% of Northern European clinical cases (Spritz et al., 1997, Halaban et al., 2000, Hutton and Spritz, 2008a). Any mutation at position 373 could result in disruption of the N-x-S/T sequence motif, which eliminates N-linked glycosylation at N-371, and produces a protein post-translationally modified with six glycans. Our most recent study with deglycosylated mutants of the tyrosinase intra-melanosomal domain, where asparagines in the glycosylation sites were replaced with aspartic acid residues, shows that deglycosylation of position 371 is critical for protein stability and activity. Specific L-DOPA activity shows moderate decrease for mutants with two glycans at positions N161 and N371 or with only one glycan at N371, and dramatic change for the fully deglycosylated protein. Moreover, the final protein purity was gradually decreased for 5, 6, and 7 N-deglycosylated mutants, compared to that of the hTyrCtr (unpublished data). This agrees with cell culture studies of T373K, which demonstrates this mutant is misfolded and retained in the ER (Halaban et al., 2000). The additional effect of T373K mutation could also be due to introduction of a positive charge close to the CuB binding region. This may inactivate the enzyme by altering the location of the histidines and/or alter the local secondary structure (Oetting and King, 1992, Tsai et al., 1999, Halaban et al., 2000).
Another common OCA1A mutant causing albinism among Korean and Japanese individuals is R77Q (c.230G>A, exon 1) (Park et al., 1996, Park et al., 1997, Tomita et al., 2000, Simeonov et al., 2013). Other mutants at position 77 also cause OCA1 (R77W) or OCA1B (R77G) forms of albinism in humans (https://portal.biobase-international.com/hgmd). Moreover, the Tyrc-2J/c-2J mouse, a model of OCA1A, is phenotypically albino due to a mutation in the Tyr gene (c.G291T, p.R77L) (Onojafe et al., 2011). In addition, in silico modeling performed in this work suggests that the R77L mutation causes a destabilizing structural change. Sterically, the human R77Q mutation could be a subject of stronger change, which might disrupt the tyrosinase catalytic activity (Shibahara et al., 1990, Spritz et al., 1997).
In our in vitro study, with recombinant T373K and R77Q mutant variants, we observed low molecular weight bands reacting with T311 antibody (Figure 2) indicating protein processing suggestive of structural perturbations. Currently, we could not provide direct evidence, we suggest OCA1A mutants were not properly folded. This agrees with previously published data from cell culture, which shows OCA1A-related mutants (possibly misfolded) are retained in the ER, bound to calnexin and calreticulin, then degraded and, thus, not released to the melanosomes, the final destination of tyrosinase (King et al., 1991, Tsai et al., 1999, Halaban et al., 2000, Berson et al., 2000).
OCA1B mutations located in exon 4, R402Q (c.1205G>A), R422W (c.1264C>T), R422Q (c.1265G>A), and P406L (c.1217C>T), display temperature-sensitive phenotypes, where the immature glycoproteins are degraded more quickly at 37˚C than at 32˚C. In cultured human melanocytes at 32˚C the enzyme is not degraded, but translocated as normal to the melanosomes (Halaban et al., 1988, Tripathi et al., 1992b, Halaban et al., 2000, Jagirdar et al., 2014). All these mutations are located in the CuB binding site and may affect the geometry of the substrate binding site or the interaction of the substrate with the binuclear copper site, resulting in reduced tyrosinase activity (Giebel et al., 1991, Oetting and King, 1992). From our results, the mutants identified in OCA1B show protein expression levels and yields similar to that of the wild type but specific activities were generally lower than the wild type protein (Table 1). Furthermore, while the hTyrCtr and R422W proteins exhibited similar free energy changes (ΔG), the other OCA1B-related mutant variants R402Q, R422Q, and P406L were less stable (Table 2). It is interesting that OCA1B-causing mutations inhibit the folding pathway to the lowest energy conformation thereby tyrosinase remains in a non-native conformation.
Mutations that change protein stability and activity are spatially close; as such, residues 402, 406, and 422 are all in the loop connecting the helix H4 from the 4-helical bundle and the protein C-terminus (Supplementary Materials, Figure S6). Genetic changes in this loop may play a role in the thermal sensitivity of human tyrosinase and possibly define the temperature-sensitive phenotypes.
There is some controversy over the mutation at position 402. As previously reported by some authors, the R402Q mutant variant is a non-pathogenic polymorphism, occuring frequently, but without strong correlation with pigmentation phenotypes among normal Caucasian individuals (Norton et al., 2007, Simeonov et al., 2013). However, R402Q is strongly associated with patients who have albinism (Chiang et al., 2009) and is not associated with autosomal recessive ocular albinism, suggesting that a causative variant may be in genetic disequilibrium with the R402Q variant (Oetting et al., 2009). On the contrary, functional in vitro studies showed the mutation at codon 402 reduces tyrosinase activity by 25% when expressed in culture HeLa cells (Tripathi et al., 1992a, Jagirdar et al., 2014). In addition, homozygous 402Q/Q human melanocytes produce significantly less tyrosinase protein, exhibit altered trafficking and glycosylation, with decreased L-DOPA activity when compare to the wild type melanocytes (Jagirdar et al., 2014). Our in vitro study of the R402Q variant showed an agreement with above results, and the reproducible decrease in enzymatic activity by 32% and corresponding decrease in protein stability (ΔΔG =0.48 kcal/mol) compared to that of the wild type (Table 1 and 2). This indicates that the R402Q might destabilize tyrosinase structure in a way similar to that of other disease-causing mutant variants from our study.
Likewise, the P406L mutation might be clinically complicated due to incorrect diagnosis in some cases as described for the TYRL pseudogene (Chaki et al., 2005). To avoid misinterpretation of the results this group designed primers for specific amplification of either TYR or TYRL locus. However, the P406L was often reported as the disease-causing mutation (Hutton and Spritz, 2008b, King et al., 2003). The proline residue has a unique stereochemistry, which restricts conformation of the protein backbone. This amino acid is often found in very tight turns and loops in protein structures and its presence has been correlated with thermostability (Matthews et al., 1987). A substitution of hydrophobic proline residue by leucine preserves a hydrophobic property in the position 406, but changes the orientation of protein backbone. This change could explain a significant destabilizing effect of the P406L mutation compared to that of the other OCA1B mutations (Table 2).
One of the limitations of our study is we used the soluble and enzymatically active domain of human tyrosinase (intra-melanosomal) as a surrogate for the full-length membrane associated protein. Although this was reasonable (and standard practice) we generated human full-length tyrosinase and showed that it had similar enzymic properties as the truncated variant. The ~ 2-fold higher activity of hTyrCtr over hTyr may be related to the fact that the latter is associated with micelles (one molecule per micelle), which could affect substrate transfer rates. It does seem reasonable, therefore, to conclude that our findings with the OCA1A variants, located in the intra-malanosomal domain, are valid.
Another issue is that in the melanosome the activity of tyrosinase might be modulated by protein-protein interactions with other proteins (Toyofuku et al., 2001). This adds an additional layer of complexity to tyrosinase and related melanogenic proteins. To resolve this issue in the future, we like to perform additional biochemical, protein-protein interactions, and functional studies on full-length tyrosinase and related proteins from the melanogenic pathway such as tyrosinase-related protein 1 (OCA3), P-protein (OCA2), among others.
In conclusion, we have shown that both OCA1A-related mutants, T373K and R77Q, although expressed in T.ni larvae, were susceptible to proteolytic degradation (a hallmark of unfolded and partially unfolded proteins) and were enzymatically inactive. This is consistent with the view that in patient’s cells OCA1A-related mutant tyrosinases are retained in the ER and degraded due to incorrect protein folding. Likewise, our results indicate that the tyrosinase catalytic activity in OCA1B- related mutants varies among the mutations and is inversely proportional to the protein stability, which is consistent with the OCA1B phenotypes variations. The biochemical and structural determinations exemplified in this study suggest how individual mutations contribute to the diverse phenotype in patients with albinism at a molecular level. Our findings may help to quantify the effect of different missense changes in OCA1B albinism. Lastly, we believe that recombinant OCA1B proteins are useful tools for identifying novel compounds that may stabilize or activate mutant tyrosinase. Such compounds may be of use in the treatment of patients with OCA1B.
Methods
Proteins expression and purification
Tyrosinase intra-melanosomal domain
Recombinant human wild type intra-melanosomal tyrosinase domain (residues 19 – 469 of the native protein), hTyrCtr and OCA1-related mutants, R422W, R402Q, R422Q, P406L, T373K, and R77Q, were expressed in baculovirus and produced in whole insect T. ni larvae at 27°C, as previously described (Dolinska et al., 2014). A 6xHis-Tag was added to the C-terminus of the protein. Protein purification was performed as follows. Frozen at −80°C, infected larvae were disrupted by Omni Tissue Homogenizer using the Hard Tissue Omni Tip™ Homogenizer Probes (Omni International, GA) in 5 x (v/w) Buffer A: 20 mM sodium phosphate, 500 mM NaCl, and 20 mM imidazole, pH 7.4, supplemented with 25 μM 1-Phenyl-2-thiourea, PTU (Sigma-Aldrich, MO), 2 mM MgCl2, 40 μg/ml DNAse I (Thermo Fisher Scientific, PA), 0.2 mg/ml lysozyme and a Complete Protease Inhibitors (Roche, USA). No additional copper was added to the buffer during the expression and purification. The homogenates were incubated on the Orbitron Rotator (Boekel Scientific, PA) at RT for 15 min and then sonicated using an Ultrasonic Processor GE130PB (Hielscher System, Germany) for 10 min. The lysates were centrifuged at 10,600 x g for 30 min at 4°C. Supernatants of hTyrCtr and mutant variants were diluted 1:1 (v/v) with binding buffer (Buffer A) and then subjected to an automated two-step purification at room temperature (RT) using ÄKTAxpress liquid chromatography system and the UNICORN 5.31 software (GE Healthcare, NJ). Crude lysate was automatically loaded on 5 ml His-Trap Crude IMAC column (GE Healthcare, NJ) equilibrated with Buffer A and eluted at flow rate of 1 ml/min into a 13-ml loop with Buffer B: 20 mM sodium phosphate, 500 mM NaCl, and 500 mM imidazole, pH 7.4. The protein from the loop was then applied on a HiPrep 26/60 Sephacryl S-300 column (GE Healthcare, NJ) pre-equilibrated with Buffer C (50 mM Tris-HCl, pH 7.4, 1mM EDTA, 150 mM NaCl, and 50 μM TCEP). Fractions containing 2 ml were collected on a 96 well plate and were analyzed by SDS-PAGE using 4–15% polyacrylamide gels (Bio-Rad, CA) stained in GelCode Blue reagent (Thermo Scientific, IL). The activity was tested by a color reaction with L-DOPA (see Tyrosinase enzymatic assays below). Protein identity was confirmed by Western blot analysis using anti-tyrosinase T311 antibody (Santa Cruz Biotechnology, CA). The proteins were concentrated using Amicon Ultra-15/10,000 NMWL centrifugal filter units (Merc Millipore, Ireland) and concentrations determined at A260/280 nm using the NanoDrop 2000c UV-Vis spectrophotometer (Thermo Scientific, DE). Tyrosinase yields for hTyrCtr and OCA1-related mutant variants in protein extracts during purification were found from SDS-PAGE gels as 100% x tyrosinase band intensity/total protein band using UN-SCAN-IT gel ™ gel analysis software (Silk Scientific, Inc., UT). Purified proteins were stored in Buffer C at −80°C until further use.
Full-length human tyrosinase
Protein (hTyr) was expressed in baculovirus, produced in whole insect T. ni larvae, homogenized in the presence of 1% Triton X-100 detergent, with supernatant prepared similar to that of described for the hTyrCtr above. The purification was performed using a Bio-Logic Duo-Flow Maximizer workstation (Bio-Rad, CA). First, the macrofiltered supernatant was loaded onto a 5 ml His-Trap FF Crude IMAC column (GE Healthcare, NJ) equilibrated preliminary with Buffer A, containing the detergent, and eluted at a flow rate of 0.5 ml/min. A gradient of 0–500 mM imidazole was applied and 2.5 ml fractions were collected. Fractions containing active hTyr were identified using a color reaction with L-DOPA. Active fractions were dialyzed overnight against 4 liters of Buffer C containing 0.1% Triton X-100. Then, the fractions were concentrated into 5 ml and were further purified by SEC using Sephacryl S200 10/60 column (GE Healthcare, NJ). Fractions of 2.5 ml were collected and monitored by SDS-PAGE, while protein identity was confirmed by Western blot analysis.
Tyrosinase enzymatic assays
Tyrosinase activities were determined spectrophotometrically (Lopez-Serrano et al., 2007, Jeong and Shim, 2004). Protein diphenol oxidase activities were measured using L-DOPA (Sigma-Aldrich, MO) as a substrate. The reaction mixture containing 3 mM L-DOPA in 10 mM sodium phosphate buffer, pH 7.4, was incubated for 30 min at 37°C and monitored by measuring the dopachrome formation at 475 nm (εdopachrome =3700 M−1 cm−1) using the SpectraMax i3 multi-mode detection platform (Molecular Devices, CA). Specific activity was determined as L-DOPA enzyme activity multiplied by the sample total volume and divided by total protein.
Kinetic parameters
The diphenol oxidase reaction rate (Vmax) was determined using L-DOPA as a substrate at concentrations of 0.094, 0.188, 0.375, 0.75, 1.5, 3, and 6 mM. All assays (in triplicate) were performed in 10 mM sodium phosphate buffer, pH 7.4 at 37°C. Absorbance was measured spectrophotometrically at 475 nm in the SpectraMax i3 multi-mode detection platform (Molecular Devices, CA) using a microtiter 96-well plate. The Michaelis-Menten constant (Km) and Vmax of proteins were calculated using SoftMax Pro software, rev. 6.5. The enyzyme turnover (kcat) was determined as a number of substrate molecules turned over per enzyme per minute, and was derived from Vmax/Et, where Et is the concentration of enzyme in mM.
Circular Dichroism (CD)
Far UV (180–260 nm) circular dichroic spectra of recombinant human hTyrCtr and OCA1B-related mutants were recorded using a Jasco 710 spectropolarimeter using a 0.1 cm path-length cell. Scans were performed in 10 mM sodium phosphate buffer, pH 7.4 at a protein concentration of 0.2 mg/ml. The reported CD spectra averaged twelve scans and corrected the values by subtracting the blank buffer. Mean residue ellipticities were expressed for all wavelengths as deg x cm2 x d.mol−1 and were calculated from the equation [θ] = θobs x M /10 x d x c, where θobs is the measured ellipticity in degrees, M, the mean residue molecular weights, d the optical path in cm and c the protein concentration in mg/ml.
Intrinsic tryptophan fluorescence
Fluorescence emission spectra of hTyrCtr and OCA1B-related mutant variants, R422W, R402Q, R422Q, and P406L (10 μM) were recorded at RT in 10 mM sodium phosphate buffer (pH 7.4) in the presence or the absence of 8 M urea. Spectra were recorded using a SpectraMax i3 multi-mode detection platform (Molecular Devices, CA). Proteins were excited at 285 nm, and the emission was recorded from 300 to 400 nm. All fluorescence spectra were corrected for the buffer baseline.
Urea-induced equilibrium unfolding/refolding
All proteins including hTyrCtr and OCA1B-related mutant variants were a subjected to equilibrium unfolding/refolding. Proteins were diluted to a final concentration of 1 μM in 10 mM phosphate buffer, pH 7.4 with urea concentrations from 0 to 8 M then incubated for 24h at RT. For equilibrium refolding experiments, 50 μM stocks of protein (final concentration 1 μM) in 8 M urea were incubated at RT for 5 h and then diluted with 10 mM phosphate buffer, pH 7.4 to 0–8 M urea and incubated for 24h at RT. Samples were excited at 285 nm and emission spectra was recorded at 300–400 nm. Both unfolding and refolding was monitored using the emission 360/320 nm ratio, which was plotted vs the urea concentration. The experimental curves were fitted with a Boltzmann function using OriginPro 2015 (Origin Lab Corporation, MA). Each unfolding/refolding curves were averaged in triplicate and normalized to show a change of unfolding fraction u from 0 to 1. The dependence of the unfolding fraction u on the urea concentration was converted to a free energy plot using the following formula : ΔG =−RT ln(u/(1−u)). Finally, the values of Gibbs free energy changes at zero urea concentration (‘physiological’ conditions) were obtained by the linear approximation of experimental points using the Origin Pro 2015 software (Origin Lab Corporation, MA).
Homology Modeling
The FASTA sequence for human tyrosinase was downloaded from the UniProtKB database (http://www.uniprot.org, Acc. # P14679) and truncated to only include residues 106–451. Homology modeling was performed using the program Yasara (http://www.yasara.org). A tyrosinase active site from Protein Model Database (model PM0079416) (Favre et al., 2014) and bacterial tyrosinase structures from the database of protein crystal structures (PDB files: 3NM8, 2AHK, 2AHL, 4J6T, and 3AWZ, http://www.rcsb.org) were used as structural templates to build an atomic model of human tyrosinase. A hydroxide ion was added in between the two copper atoms to more accurately match the PM0079416 model and depict the likelihood of a hydroxide ion present in Tyr’s met form. The OH− ion was placed as a bridge between Cu A and B. Copper A was localized to be, approximately, 2.0Å in distance from the epsilon nitrogen, Nε2, on histidine 180, 202, and 211. The same distance was determined for copper B and the Nε2 atom of histidine 363, 367, and 390. Finally, the atomic structure of human tyrosinase was equilibrated in water for 40ns using scalable molecular dynamics program NAMD (http://www.ks.uiuc.edu/Research/namd/). Finally, the obtained structure was used for modeling of the following mutant variants: T373K/N371D, P406L, R402Q, R422W, and R422Q. The tyrosinases stereochemistry was repaired and evaluated for free energy changes, ΔΔG using the FoldX suite (http://foldxsuite.crg.eu/). The structure of R77Q was not modeled because of the residues 77 was located outside of the range for our current structure.
Supplementary Material
Figure S1. Expression of hTyrCtr and OCA1-related mutants in lysates from T.ni larvae.
SDS-PAGE (Panel A) and Western blot (Panel B) showing the expression of hTyrCtr and OCA1-related mutants. From the left: L, protein ladder; C, positive control (pure hTyrCtr migrates as a broad band due to glycosylation), and total lysate of larvae expressing hTyrCtr, R422W, R402Q, R422Q, P406L, T373K, and R77Q. The table at the bottom shows the yield of each protein expressed in lysates measured from SDS-PAGE gels as 100% x tyrosinase band intensity/total protein band using N-SCAN-IT gel ™ gel analysis software.
Figure S2. Gel filtration and far-UV circular dichroism for hTyrCtr and OCA1B-related mutants.
Panel A. Size-exclusion chromatography profiles of hTyrCtr (red) and mutant variants (R422W, orange; R402Q, yellow; R422Q, green, and P406L, blue) were eluted from Superose 12 10/300 GL column as single monomeric peaks with an averaged molecular weight of 54.96 ± 1.41 kDa. The elution points of molecular weight standards (thyroglobulin 670.0 kDa, gamma globulin 155.0 kDa, ovalbumin 44.0 kDa, and myoglobin 17.0 kDa) are shown at the top for reference. Proteins concentration was monitored by absorbance at 280 nm. Panel B. CD scans (180 – 260 nm) were performed in 10 mM sodium phosphate buffer, pH 7.4 at protein concentration of 0.2 mg/ml. SEC profiles and CD spectra for hTyrCtr and OCA1B-related mutants, R422W, R402Q, R422Q, and P406L, are shown by red, orange, yellow, green, and blue lines, respectively.
Figure S3. Tryptophan residues are shown in hTyrCtr
Tryptophan (Trp) residues mapped to the human 40 ns tyrosinase protein structure modeled as described in methods section. Panel A shown the protein backbone structure (gold ribbon) and Trp residues, which are represented by orange spheres. Accessibility calculations using the same model show that 6 tryptophans are buried (residues 160, 192, 200, 220, 254, and 382) and 5 are partially exposed (residues 21, 62, 90, 177, and 218). In Panel B the protein surface is shown by the gold mesh.
Figure S4. Intrinsic tryptophan fluorescence spectra of the native, unfolded and refolded states of hTyrCtr and OCA1B-related mutants.
Native, unfolded, and refolded proteins are indicated by grey, black, and green lines, respectively. Proteins were excited at 285 nm and emission spectra recorded from 300 – 400 nm. For native and unfolded state, proteins of 1 μM were incubated in 10 mM phosphate buffer, pH 7.4, 50 μM TCEP with or without 8 M urea at RT for 24 h. For refolding, 50 μM stocks of protein in 8 M urea were incubated at RT for 5 h and then diluted with 10 mM phosphate buffer, pH 7.4 contained 0–8 M urea up to a final protein concentration of 1 μM. To reach the equilibrium, samples were incubated for at RT 24 h. The inserted table shows the maximum wavelengths (λmax) of hTyrCtr and OCA1B-related mutants under the native, unfolded, and refolded states.
Figure S5. Maximum wavelength relative shift after unfolding/refolding shown for hTyrCtr and OCA1B-related mutants.
Maximum wavelength relative shift of tryptophan fluorescence emission spectra of unfolded (red bars) and refolded (blue bars) proteins. Proteins (10 μM) in 10 mM phosphate buffer, pH 7.0, 50 μM TCEP were excited at 285 nm and emission spectra were recorded from 300 – 400 nm. Refolding yield in percent is indicated.
Figure S6. OCA1B-related mutations co-localized within the same structural element of the tyrosinase structure.
The 4-helical bundle formed by helices H1–H4 is shown by baby blue ribbon. Two copper atoms depicted as orange spheres are coordinated by six histidines in the center of the active site. Residues R402, P406, and R422 (red) are located on the same loop (beige), which connects the helix H4 and the protein C-terminus. In the full-length tyrosinase is connected to the trans-membrane helix (TM).
Significance.
Very little is known how individual mutations contribute to the diverse pigmentation phenotype in patients with OCA1 albinism at the molecular level. Our in vitro experiments with purified recombinant human tyrosinase mutants corresponding to OCA1A indicate they are unstable and enzymatically inactive. In contrast, OCA1B mutants are more stable and active in varying degrees. These results suggest a direct link between protein stability and loss of pigmentation in OC1A. This opens the possibility of high-throughput screens to identify novel drug compounds, which modulate protein folding and stability, resulting in recovery of tyrosinase activity and pigmentation in albinism patients.
Abbreviations
- OCA1
oculocutaneous albinism Type 1 genetic disorder
- OCA1A
a Type 1A disorder with a complete absence of tyrosinase activity and lack of melanin production
- OCA1B
a Type 1B disorder with residual tyrosinase activity
- hTyrCtr and hTyr
intra-melanosomal domain and full-length human tyrosinase, respectively
- IMAC
immobilized metal affinity chromatography
- SEC
size-exclusion chromatography
- CD
circular dichroism
- L-DOPA
L-3,4-dihydroxyphenylalanine
- PTU
1-Phenyl-2-thiourea
- RT
room temperature
References
- AMMANN S, SCHULZ A, KRAGELOH-MANN I, DIECKMANN NM, NIETHAMMER K, FUCHS S, ECKL KM, PLANK R, WERNER R, ALTMULLER J, THIELE H, NURNBERG P, BANK J, STRAUSS A, VON BERNUTH H, ZUR STADT U, GRIEVE S, GRIFFITHS GM, LEHMBERG K, HENNIES HC, EHL S. Mutations in AP3D1 associated with immunodeficiency and seizures define a new type of Hermansky-Pudlak syndrome. Blood. 2016;127:997–1006. doi: 10.1182/blood-2015-09-671636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANIKSTER Y, HUIZING M, WHITE J, SHEVCHENKO YO, FITZPATRICK DL, TOUCHMAN JW, COMPTON JG, BALE SJ, SWANK RT, GAHL WA, TORO JR. Mutation of a new gene causes a unique form of Hermansky-Pudlak syndrome in a genetic isolate of central Puerto Rico. Nat Genet. 2001;28:376–80. doi: 10.1038/ng576. [DOI] [PubMed] [Google Scholar]
- BERSON JF, FRANK DW, CALVO PA, BIELER BM, MARKS MS. A common temperature-sensitive allelic form of human tyrosinase is retained in the endoplasmic reticulum at the nonpermissive temperature. J Biol Chem. 2000;275:12281–9. doi: 10.1074/jbc.275.16.12281. [DOI] [PubMed] [Google Scholar]
- BIJELIC A, PRETZLER M, MOLITOR C, ZEKIRI F, ROMPEL A. The Structure of a Plant Tyrosinase from Walnut Leaves Reveals the Importance of “Substrate-Guiding Residues” for Enzymatic Specificity. Angew Chem Int Ed Engl. 2015;54:14677–80. doi: 10.1002/anie.201506994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOISSY RE, ZHAO H, OETTING WS, AUSTIN LM, WILDENBERG SC, BOISSY YL, ZHAO Y, STURM RA, HEARING VJ, KING RA, NORDLUND JJ. Mutation in and lack of expression of tyrosinase-related protein-1 (TRP-1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as “OCA3”. Am J Hum Genet. 1996;58:1145–56. [PMC free article] [PubMed] [Google Scholar]
- CHAKI M, MUKHOPADHYAY A, RAY K. Determination of variants in the 3′-region of the tyrosinase gene requires locus specific amplification. Hum Mutat. 2005;26:53–8. doi: 10.1002/humu.20171. [DOI] [PubMed] [Google Scholar]
- CHIANG PW, SPECTOR E, TSAI AC. Oculocutaneous albinism spectrum. Am J Med Genet A. 2009;149A:1590–1. doi: 10.1002/ajmg.a.32939. [DOI] [PubMed] [Google Scholar]
- CULLINANE AR, CURRY JA, CARMONA-RIVERA C, SUMMERS CG, CICCONE C, CARDILLO ND, DORWARD H, HESS RA, WHITE JG, ADAMS D, HUIZING M, GAHL WA. A BLOC-1 mutation screen reveals that PLDN is mutated in Hermansky-Pudlak Syndrome type 9. Am J Hum Genet. 2011;88:778–87. doi: 10.1016/j.ajhg.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- DOLINSKA MB, KOVALEVA E, BACKLUND P, WINGFIELD PT, BROOKS BP, SERGEEV YV. Albinism-causing mutations in recombinant human tyrosinase alter intrinsic enzymatic activity. PLoS One. 2014;9:e84494. doi: 10.1371/journal.pone.0084494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAVRE E, DAINA A, CARRUPT PA, NURISSO A. Modeling the met form of human tyrosinase: a refined and hydrated pocket for antagonist design. Chem Biol Drug Des. 2014;84:206–15. doi: 10.1111/cbdd.12306. [DOI] [PubMed] [Google Scholar]
- GIEBEL LB, TRIPATHI RK, KING RA, SPRITZ RA. A tyrosinase gene missense mutation in temperature-sensitive type I oculocutaneous albinism. A human homologue to the Siamese cat and the Himalayan mouse. J Clin Invest. 1991;87:1119–22. doi: 10.1172/JCI115075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRONSKOV K, DOOLEY CM, OSTERGAARD E, KELSH RN, HANSEN L, LEVESQUE MP, VILHELMSEN K, MOLLGARD K, STEMPLE DL, ROSENBERG T. Mutations in c10orf11, a melanocyte-differentiation gene, cause autosomal-recessive albinism. Am J Hum Genet. 2013;92:415–21. doi: 10.1016/j.ajhg.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRONSKOV K, EK J, BRONDUM-NIELSEN K. Oculocutaneous albinism. Orphanet J Rare Dis. 2007;2:43. doi: 10.1186/1750-1172-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALABAN R, CHENG E, SVEDINE S, ARON R, HEBERT DN. Proper folding and endoplasmic reticulum to golgi transport of tyrosinase are induced by its substrates, DOPA and tyrosine. J Biol Chem. 2001;276:11933–8. doi: 10.1074/jbc.M008703200. [DOI] [PubMed] [Google Scholar]
- HALABAN R, MOELLMANN G, TAMURA A, KWON BS, KUKLINSKA E, POMERANTZ SH, LERNER AB. Tyrosinases of murine melanocytes with mutations at the albino locus. Proc Natl Acad Sci U S A. 1988;85:7241–5. doi: 10.1073/pnas.85.19.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALABAN R, SVEDINE S, CHENG E, SMICUN Y, ARON R, HEBERT DN. Endoplasmic reticulum retention is a common defect associated with tyrosinase-negative albinism. Proc Natl Acad Sci U S A. 2000;97:5889–94. doi: 10.1073/pnas.97.11.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEARING VJ, JR, EKEL TM, MONTAGUE PM, HEARING ED, NICHOLSON JM. Mammalian tyrosinase: isolation by a simple new procedure and characterization of its steric requirements for cofactor activity. Arch Biochem Biophys. 1978;185:407–18. doi: 10.1016/0003-9861(78)90183-2. [DOI] [PubMed] [Google Scholar]
- HUIZING M, PEDERSON B, HESS RA, GRIFFIN A, HELIP-WOOLEY A, WESTBROEK W, DORWARD H, O’BRIEN KJ, GOLAS G, TSILOU E, WHITE JG, GAHL WA. Clinical and cellular characterisation of Hermansky-Pudlak syndrome type 6. J Med Genet. 2009;46:803–10. doi: 10.1136/jmg.2008.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUTTON SM, SPRITZ RA. Comprehensive analysis of oculocutaneous albinism among non-Hispanic caucasians shows that OCA1 is the most prevalent OCA type. J Invest Dermatol. 2008a;128:2442–50. doi: 10.1038/jid.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUTTON SM, SPRITZ RA. A comprehensive genetic study of autosomal recessive ocular albinism in Caucasian patients. Invest Ophthalmol Vis Sci. 2008b;49:868–72. doi: 10.1167/iovs.07-0791. [DOI] [PubMed] [Google Scholar]
- IMOKAWA G, MISHIMA Y. Functional analysis of tyrosinase isozymes of cultured malignant melanoma cells during the recovery period following interrupted melanogenesis induced by glycosylation inhibitors. J Invest Dermatol. 1984;83:196–201. doi: 10.1111/1523-1747.ep12263562. [DOI] [PubMed] [Google Scholar]
- JAGIRDAR K, SMIT DJ, AINGER SA, LEE KJ, BROWN DL, CHAPMAN B, ZHEN ZHAO Z, MONTGOMERY GW, MARTIN NG, STOW JL, DUFFY DL, STURM RA. Molecular analysis of common polymorphisms within the human Tyrosinase locus and genetic association with pigmentation traits. Pigment Cell Melanoma Res. 2014;27:552–64. doi: 10.1111/pcmr.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JEONG CH, SHIM KH. Tyrosinase inhibitor isolated from the leaves of Zanthoxylum piperitum. Biosci Biotechnol Biochem. 2004;68:1984–7. doi: 10.1271/bbb.68.1984. [DOI] [PubMed] [Google Scholar]
- JUNG J, BOHN G, ALLROTH A, BOZTUG K, BRANDES G, SANDROCK I, SCHAFFER AA, RATHINAM C, KOLLNER I, BEGER C, SCHILKE R, WELTE K, GRIMBACHER B, KLEIN C. Identification of a homozygous deletion in the AP3B1 gene causing Hermansky-Pudlak syndrome, type 2. Blood. 2006;108:362–9. doi: 10.1182/blood-2005-11-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KING RA, MENTINK MM, OETTING WS. Non-random distribution of missense mutations within the human tyrosinase gene in type I (tyrosinase-related) oculocutaneous albinism. Mol Biol Med. 1991;8:19–29. [PubMed] [Google Scholar]
- KING RA, OLDS DP, WITKOP CJ. Characterization of human hairbulb tyrosinase: properties of normal and albino enzyme. J Invest Dermatol. 1978;71:136–9. doi: 10.1111/1523-1747.ep12546765. [DOI] [PubMed] [Google Scholar]
- KING RA, PIETSCH J, FRYER JP, SAVAGE S, BROTT MJ, RUSSELL-EGGITT I, SUMMERS CG, OETTING WS. Tyrosinase gene mutations in oculocutaneous albinism 1 (OCA1): definition of the phenotype. Hum Genet. 2003;113:502–13. doi: 10.1007/s00439-003-0998-1. [DOI] [PubMed] [Google Scholar]
- KING RA, WIRTSCHAFTER JD, OLDS DP, BRUMBAUGH J. Minimal pigment: a new type of oculocutaneous albinism. Clin Genet. 1986;29:42–50. doi: 10.1111/j.1399-0004.1986.tb00769.x. [DOI] [PubMed] [Google Scholar]
- KOHASHI PY, KUMAGAI T, MATOBA Y, YAMAMOTO A, MARUYAMA M, SUGIYAMA M. An efficient method for the overexpression and purification of active tyrosinase from Streptomyces castaneoglobisporus. Protein Expr Purif. 2004;34:202–7. doi: 10.1016/j.pep.2003.11.015. [DOI] [PubMed] [Google Scholar]
- LI W, ZHANG Q, OISO N, NOVAK EK, GAUTAM R, O’BRIEN EP, TINSLEY CL, BLAKE DJ, SPRITZ RA, COPELAND NG, JENKINS NA, AMATO D, ROE BA, STARCEVIC M, DELL’ANGELICA EC, ELLIOTT RW, MISHRA V, KINGSMORE SF, PAYLOR RE, SWANK RT. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nat Genet. 2003;35:84–9. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOPEZ-SERRANO D, SOLANO F, SANCHEZ-AMAT A. Involvement of a novel copper chaperone in tyrosinase activity and melanin synthesis in Marinomonas mediterranea. Microbiology. 2007;153:2241–9. doi: 10.1099/mic.0.2007/006833-0. [DOI] [PubMed] [Google Scholar]
- MANGA P, KROMBERG JG, BOX NF, STURM RA, JENKINS T, RAMSAY M. Rufous oculocutaneous albinism in southern African Blacks is caused by mutations in the TYRP1 gene. Am J Hum Genet. 1997;61:1095–101. doi: 10.1086/301603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATTHEWS BW, NICHOLSON H, BECKTEL WJ. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci U S A. 1987;84:6663–7. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONTOLIU L, GRONSKOV K, WEI AH, MARTINEZ-GARCIA M, FERNANDEZ A, ARVEILER B, MORICE-PICARD F, RIAZUDDIN S, SUZUKI T, AHMED ZM, ROSENBERG T, LI W. Increasing the complexity: new genes and new types of albinism. Pigment Cell Melanoma Res. 2014;27:11–8. doi: 10.1111/pcmr.12167. [DOI] [PubMed] [Google Scholar]
- MORGAN NV, PASHA S, JOHNSON CA, AINSWORTH JR, EADY RA, DAWOOD B, MCKEOWN C, TREMBATH RC, WILDE J, WATSON SP, MAHER ER. A germline mutation in BLOC1S3/reduced pigmentation causes a novel variant of Hermansky-Pudlak syndrome (HPS8) Am J Hum Genet. 2006;78:160–6. doi: 10.1086/499338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEWTON JM, COHEN-BARAK O, HAGIWARA N, GARDNER JM, DAVISSON MT, KING RA, BRILLIANT MH. Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet. 2001;69:981–8. doi: 10.1086/324340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORTON HL, KITTLES RA, PARRA E, MCKEIGUE P, MAO X, CHENG K, CANFIELD VA, BRADLEY DG, MCEVOY B, SHRIVER MD. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 2007;24:710–22. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- OETTING WS. The tyrosinase gene and oculocutaneous albinism type 1 (OCA1): A model for understanding the molecular biology of melanin formation. Pigment Cell Res. 2000;13:320–5. doi: 10.1034/j.1600-0749.2000.130503.x. [DOI] [PubMed] [Google Scholar]
- OETTING WS, KING RA. Analysis of mutations in the copper B binding region associated with type I (tyrosinase-related) oculocutaneous albinism. Pigment Cell Res. 1992;5:274–8. doi: 10.1111/j.1600-0749.1992.tb00549.x. [DOI] [PubMed] [Google Scholar]
- OETTING WS, PIETSCH J, BROTT MJ, SAVAGE S, FRYER JP, SUMMERS CG, KING RA. The R402Q tyrosinase variant does not cause autosomal recessive ocular albinism. Am J Med Genet A. 2009;149A:466–9. doi: 10.1002/ajmg.a.32654. [DOI] [PubMed] [Google Scholar]
- OH J, HO L, ALA-MELLO S, AMATO D, ARMSTRONG L, BELLUCCI S, CARAKUSHANSKY G, ELLIS JP, FONG CT, GREEN JS, HEON E, LEGIUS E, LEVIN AV, NIEUWENHUIS HK, PINCKERS A, TAMURA N, WHITEFORD ML, YAMASAKI H, SPRITZ RA. Mutation analysis of patients with Hermansky-Pudlak syndrome: a frameshift hot spot in the HPS gene and apparent locus heterogeneity. Am J Hum Genet. 1998;62:593–8. doi: 10.1086/301757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ONOJAFE IF, ADAMS DR, SIMEONOV DR, ZHANG J, CHAN CC, BERNARDINI IM, SERGEEV YV, DOLINSKA MB, ALUR RP, BRILLIANT MH, GAHL WA, BROOKS BP. Nitisinone improves eye and skin pigmentation defects in a mouse model of oculocutaneous albinism. J Clin Invest. 2011;121:3914–23. doi: 10.1172/JCI59372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK KC, PARK SK, LEE YS, YOUN SW, PARK BS, KIM KH, LEE ST. Mutations of the tyrosinase gene in three Korean patients with type I oculocutaneous albinism. Jpn J Hum Genet. 1996;41:299–305. doi: 10.1007/BF01913172. [DOI] [PubMed] [Google Scholar]
- PARK SK, LEE KH, PARK KC, LEE JS, SPRITZ RA, LEE ST. Prevalent and novel mutations of the tyrosinase gene in Korean patients with tyrosinase-deficient oculocutaneous albinism. Mol Cells. 1997;7:187–91. [PubMed] [Google Scholar]
- POPESCU CI, PADURARU C, DWEK RA, PETRESCU SM. Soluble tyrosinase is an endoplasmic reticulum (ER)-associated degradation substrate retained in the ER by calreticulin and BiP/GRP78 and not calnexin. J Biol Chem. 2005;280:13833–40. doi: 10.1074/jbc.M413087200. [DOI] [PubMed] [Google Scholar]
- RILEY PA. The great DOPA mystery: the source and significance of DOPA in phase I melanogenesis. Cell Mol Biol (Noisy-le-grand) 1999;45:951–60. [PubMed] [Google Scholar]
- SCHIAFFINO MV, TACCHETTI C. The ocular albinism type 1 (OA1) protein and the evidence for an intracellular signal transduction system involved in melanosome biogenesis. Pigment Cell Res. 2005;18:227–33. doi: 10.1111/j.1600-0749.2005.00240.x. [DOI] [PubMed] [Google Scholar]
- SHIBAHARA S, OKINAGA S, TOMITA Y, TAKEDA A, YAMAMOTO H, SATO M, TAKEUCHI T. A point mutation in the tyrosinase gene of BALB/c albino mouse causing the cysteine----serine substitution at position 85. Eur J Biochem. 1990;189:455–61. doi: 10.1111/j.1432-1033.1990.tb15510.x. [DOI] [PubMed] [Google Scholar]
- SIMEONOV DR, WANG X, WANG C, SERGEEV Y, DOLINSKA M, BOWER M, FISCHER R, WINER D, DUBROVSKY G, BALOG JZ, HUIZING M, HART R, ZEIN WM, GAHL WA, BROOKS BP, ADAMS DR. DNA variations in oculocutaneous albinism: an updated mutation list and current outstanding issues in molecular diagnostics. Hum Mutat. 2013;34:827–35. doi: 10.1002/humu.22315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPRITZ RA, HO L, FURUMURA M, HEARING VJ., JR Mutational analysis of copper binding by human tyrosinase. J Invest Dermatol. 1997;109:207–12. doi: 10.1111/1523-1747.ep12319351. [DOI] [PubMed] [Google Scholar]
- SUZUKI T, LI W, ZHANG Q, KARIM A, NOVAK EK, SVIDERSKAYA EV, HILL SP, BENNETT DC, LEVIN AV, NIEUWENHUIS HK, FONG CT, CASTELLAN C, MITERSKI B, SWANK RT, SPRITZ RA. Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat Genet. 2002;30:321–4. doi: 10.1038/ng835. [DOI] [PubMed] [Google Scholar]
- TOMITA Y, MIYAMURA Y, KONO M, NAKAMURA R, MATSUNAGA J. Molecular bases of congenital hypopigmentary disorders in humans and oculocutaneous albinism 1 in Japan. Pigment Cell Res. 2000;13(Suppl 8):130–4. doi: 10.1034/j.1600-0749.13.s8.23.x. [DOI] [PubMed] [Google Scholar]
- TOYOFUKU K, WADA I, VALENCIA JC, KUSHIMOTO T, FERRANS VJ, HEARING VJ. Oculocutaneous albinism types 1 and 3 are ER retention diseases: mutation of tyrosinase or Tyrp1 can affect the processing of both mutant and wild-type proteins. FASEB J. 2001;15:2149–61. doi: 10.1096/fj.01-0216com. [DOI] [PubMed] [Google Scholar]
- TRIPATHI RK, HEARING VJ, URABE K, AROCA P, SPRITZ RA. Mutational mapping of the catalytic activities of human tyrosinase. J Biol Chem. 1992a;267:23707–12. [PubMed] [Google Scholar]
- TRIPATHI RK, STRUNK KM, GIEBEL LB, WELEBER RG, SPRITZ RA. Tyrosinase gene mutations in type I (tyrosinase-deficient) oculocutaneous albinism define two clusters of missense substitutions. Am J Med Genet. 1992b;43:865–71. doi: 10.1002/ajmg.1320430523. [DOI] [PubMed] [Google Scholar]
- TSAI CH, TSAI FJ, WU JY, LIN SP, CHANG JG, YANG CF, LEE CC. Insertion/deletion mutations of type I oculocutaneous albinism in chinese patients from Taiwan. Hum Mutat. 1999;14:542. doi: 10.1002/(SICI)1098-1004(199912)14:6<542::AID-HUMU14>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- WANG N, HEBERT DN. Tyrosinase maturation through the mammalian secretory pathway: bringing color to life. Pigment Cell Res. 2006;19:3–18. doi: 10.1111/j.1600-0749.2005.00288.x. [DOI] [PubMed] [Google Scholar]
- WEI AH, ZANG DJ, ZHANG Z, LIU XZ, HE X, YANG L, WANG Y, ZHOU ZY, ZHANG MR, DAI LL, YANG XM, LI W. Exome sequencing identifies SLC24A5 as a candidate gene for nonsyndromic oculocutaneous albinism. J Invest Dermatol. 2013;133:1834–40. doi: 10.1038/jid.2013.49. [DOI] [PubMed] [Google Scholar]
- ZHANG Q, ZHAO B, LI W, OISO N, NOVAK EK, RUSINIAK ME, GAUTAM R, CHINTALA S, O’BRIEN EP, ZHANG Y, ROE BA, ELLIOTT RW, EICHER EM, LIANG P, KRATZ C, LEGIUS E, SPRITZ RA, O’SULLIVAN TN, COPELAND NG, JENKINS NA, SWANK RT. Ru2 and Ru encode mouse orthologs of the genes mutated in human Hermansky-Pudlak syndrome types 5 and 6. Nat Genet. 2003;33:145–53. doi: 10.1038/ng1087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Expression of hTyrCtr and OCA1-related mutants in lysates from T.ni larvae.
SDS-PAGE (Panel A) and Western blot (Panel B) showing the expression of hTyrCtr and OCA1-related mutants. From the left: L, protein ladder; C, positive control (pure hTyrCtr migrates as a broad band due to glycosylation), and total lysate of larvae expressing hTyrCtr, R422W, R402Q, R422Q, P406L, T373K, and R77Q. The table at the bottom shows the yield of each protein expressed in lysates measured from SDS-PAGE gels as 100% x tyrosinase band intensity/total protein band using N-SCAN-IT gel ™ gel analysis software.
Figure S2. Gel filtration and far-UV circular dichroism for hTyrCtr and OCA1B-related mutants.
Panel A. Size-exclusion chromatography profiles of hTyrCtr (red) and mutant variants (R422W, orange; R402Q, yellow; R422Q, green, and P406L, blue) were eluted from Superose 12 10/300 GL column as single monomeric peaks with an averaged molecular weight of 54.96 ± 1.41 kDa. The elution points of molecular weight standards (thyroglobulin 670.0 kDa, gamma globulin 155.0 kDa, ovalbumin 44.0 kDa, and myoglobin 17.0 kDa) are shown at the top for reference. Proteins concentration was monitored by absorbance at 280 nm. Panel B. CD scans (180 – 260 nm) were performed in 10 mM sodium phosphate buffer, pH 7.4 at protein concentration of 0.2 mg/ml. SEC profiles and CD spectra for hTyrCtr and OCA1B-related mutants, R422W, R402Q, R422Q, and P406L, are shown by red, orange, yellow, green, and blue lines, respectively.
Figure S3. Tryptophan residues are shown in hTyrCtr
Tryptophan (Trp) residues mapped to the human 40 ns tyrosinase protein structure modeled as described in methods section. Panel A shown the protein backbone structure (gold ribbon) and Trp residues, which are represented by orange spheres. Accessibility calculations using the same model show that 6 tryptophans are buried (residues 160, 192, 200, 220, 254, and 382) and 5 are partially exposed (residues 21, 62, 90, 177, and 218). In Panel B the protein surface is shown by the gold mesh.
Figure S4. Intrinsic tryptophan fluorescence spectra of the native, unfolded and refolded states of hTyrCtr and OCA1B-related mutants.
Native, unfolded, and refolded proteins are indicated by grey, black, and green lines, respectively. Proteins were excited at 285 nm and emission spectra recorded from 300 – 400 nm. For native and unfolded state, proteins of 1 μM were incubated in 10 mM phosphate buffer, pH 7.4, 50 μM TCEP with or without 8 M urea at RT for 24 h. For refolding, 50 μM stocks of protein in 8 M urea were incubated at RT for 5 h and then diluted with 10 mM phosphate buffer, pH 7.4 contained 0–8 M urea up to a final protein concentration of 1 μM. To reach the equilibrium, samples were incubated for at RT 24 h. The inserted table shows the maximum wavelengths (λmax) of hTyrCtr and OCA1B-related mutants under the native, unfolded, and refolded states.
Figure S5. Maximum wavelength relative shift after unfolding/refolding shown for hTyrCtr and OCA1B-related mutants.
Maximum wavelength relative shift of tryptophan fluorescence emission spectra of unfolded (red bars) and refolded (blue bars) proteins. Proteins (10 μM) in 10 mM phosphate buffer, pH 7.0, 50 μM TCEP were excited at 285 nm and emission spectra were recorded from 300 – 400 nm. Refolding yield in percent is indicated.
Figure S6. OCA1B-related mutations co-localized within the same structural element of the tyrosinase structure.
The 4-helical bundle formed by helices H1–H4 is shown by baby blue ribbon. Two copper atoms depicted as orange spheres are coordinated by six histidines in the center of the active site. Residues R402, P406, and R422 (red) are located on the same loop (beige), which connects the helix H4 and the protein C-terminus. In the full-length tyrosinase is connected to the trans-membrane helix (TM).