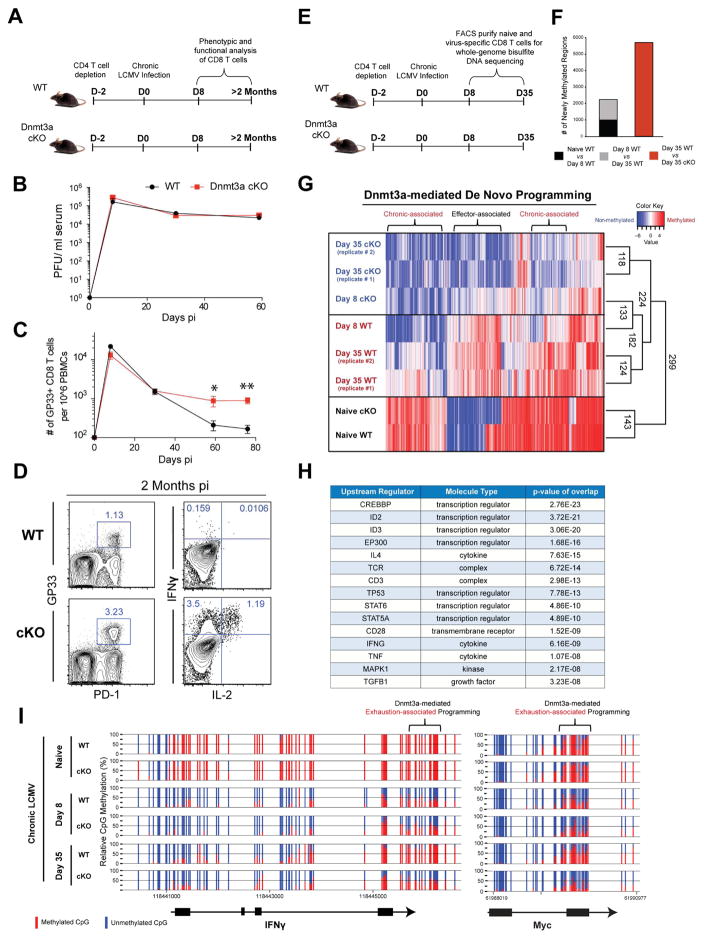
Figure 1. Whole-Genome methylation profiling of LCMV-specific WT and Dnmt3a-Deficient CD8 T Cells.
(A) Experimental setup; CD4 T cells were depleted in WT and Dnmt3a-cKO mice prior to infection with LCMV clone 13.
(B) Longitudinal summary graph of WT and Dnmt3a cKO mice serum viral titers during chronic LCMV infection.
(C) Summary graph of gp33-specific CD8 T cell quantity in the peripheral blood of WT and cKO mice during chronic LCMV infection.
(D) Representative FACS analysis of PD-1 expression on gp33-specific CD8 T cells from the spleens of WT and cKO mice after 2 months of chronic infection, gated on CD8+ T cells. Representative intracellular FACS analysis of IFNγ and IL-2 expression from gp33 peptide-stimulated CD44hi CD8 T cell splenocytes of chronically infected WT and cKO mice.
(E) Experimental setup for performing whole-genome bisulfite sequencing (WGBS) methylation analysis of virus-specific CD8 T cells.
(F) The number of newly methylated regions in gp33-specific effector or exhausted WT CD8 T cells.
(G) Heat map showing cluster analysis of the top 3000 differentially methylated regions (DMRs) among naïve and LCMV-specific WT and cKO CD8 T cells at the effector and chronic stages of the immune response. Color intensity scales from red (methylated region) to blue (non-methylated region).
(H) Summary table of ingenuity pathway analysis showing putative upstream regulators for Dnmt3a-targeted genes.
(I) Nucleotide-resolution methylation profiling of the IFNγ and Myc loci in naïve and LCMV-specific CD8 T cells. Vertical blue and red lines indicate CpG positions in the loci. The ratio of blue to red indicates the percentage of unmethylated versus methylated reads, respectively, in the WGBS.
N= 3–5 mice per group of two or more independent experiments. Error bars indicate SEM.