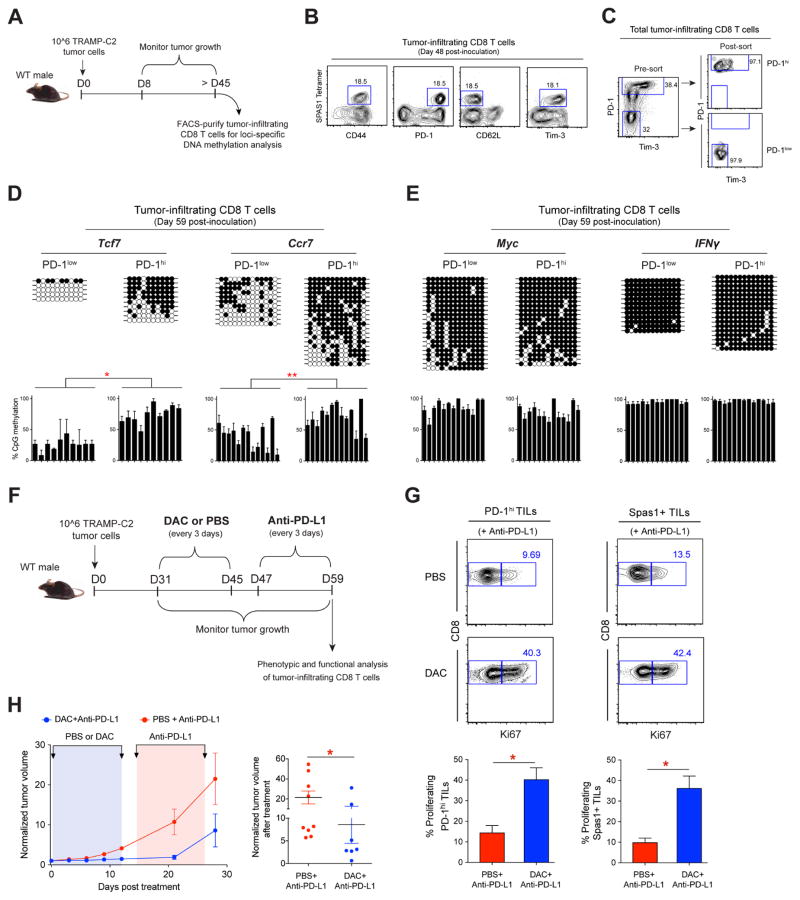
Figure 7. Tumor-Infiltrating CD8 T Cells Acquire Exhaustion-Associated DNA Methylation Programs.
(A) Experimental setup for generating TRAMP-C2 tumor-bearing mice and loci-specific DNA methylation analysis of tumor-infiltrating CD8 T cells (TILs).
(B) Representative FACS analysis of CD44, PD-1, CD62L, and Tim-3 expression on Tumor-specific Spas1+ TILs harvested after ≥ 45 days post-tumor inoculation.
(C) Representative FACS analysis of PD-1 and Tim-3 expression on total TILs and post-sort purity check of FACS-purified PD-1hi and PD-1low TILs populations.
(D) Loci-specific bisulfite sequencing methylation analysis and summary graphs of individual CpG sites in the Tcf7, Ccr7, (E) Myc and IFNγ loci among PD-1low and PD-1hi populations of TILs. Horizontal lines represent individual sequenced clones from the pool of FACS-purified TILs. Filled circles = methylated cytosine; open circles = nonmethylated cytosine.
(F) Experimental setup of sequential decitabine and anti-PD-L1 treatment of TRAMP-C2 tumor-bearing mice starting at ≥ 1 month post-inoculation.
(G) Representative FACS analysis and summary graphs of Ki67 levels among PD-1hi CD8+ TILs and tumor antigen-specific (Spas1+) TILs after mono PD-1 blockade or sequentially combined decitabine and anti-PD-L1 treatments.
(H) Longitudinal measurement of tumor growth in WT mice receiving mono anti-PD-L1 (red), or sequentially combined decitabine and anti-PD-L1 (blue) treatments. Summary graph of normalized tumor volume after mono anti-PD-L1 (red) or combined decitabine and anti-PD-L1 (blue) treatments.
N= 3–5 mice per group of two independent experiments. Error bars indicate SEM.