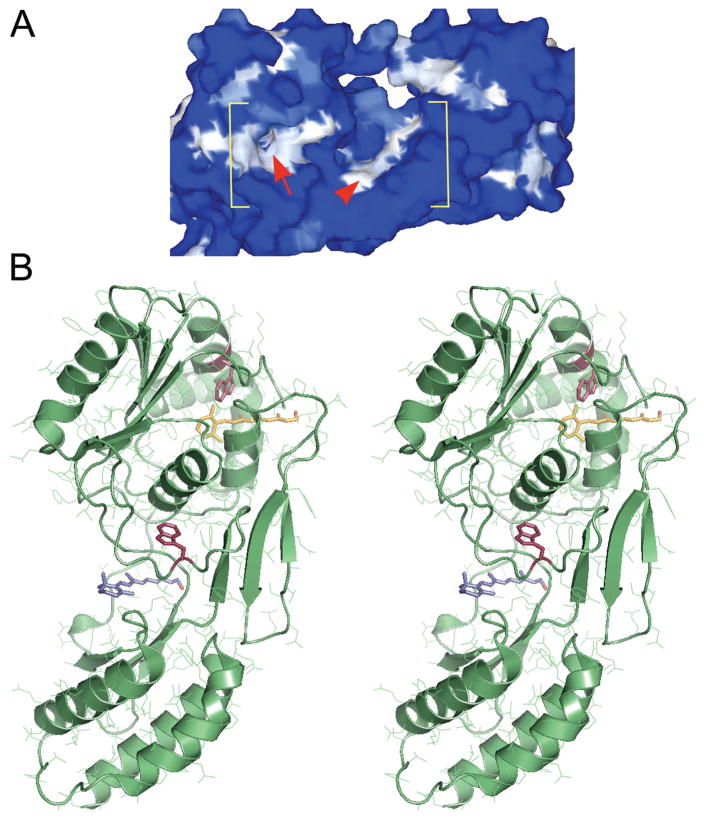
Figure 1.
Two putative retinol-binding sites within the second module of Xenopus IRBP (X2IRBP). (A) Hydrophobicity surface representation, where white represents high hydrophobicity, and blue represents high hydrophilicity. Within the bracketed area, the arrowhead identifies a shallow hydrophobic cleft. Arrow: an opening located in a hydrophobic patch that leads to a hydrophobic cavity. Modified with permission from Loew A, Gonzalez-Fernandez F. Crystal structure of the functional unit of interphotoreceptor retinoid binding protein. Structure. 2002;10(1):43–49. © Cell Press. (B) Stereoimage of computer docking of all-trans retinol in each of the two possible hydrophobic ligand-binding domains. We have previously shown that retinol-binding quenches tryptophan fluorescence in X2IRBP (Gonzalez-Fernandez et al.38). There are two tryptophan residues within X2IRBP that could support this quenching. Each of these tryptophans (red) is located in one of the putative ligand-binding domains. W450 is located in the hydrophobic cavity containing a molecule of retinol (yellow); W587 is located in the more shallow hydrophobic cleft containing a molecule of retinol (blue). Image modified with permission from Gonzalez-Fernandez F, Ghosh D. Focus on molecules: interphotoreceptor retinoid-binding protein (IRBP). Exp Eye Res. 2006;86:169–170. © Elsevier. The image may be viewed in 3-D without specialized stereo glasses. Suggestions for viewing molecular stereo images are available at http://spdbv.vital-it.ch/TheMolecularLevel/0Help/StereoView.html.