Figure 5.
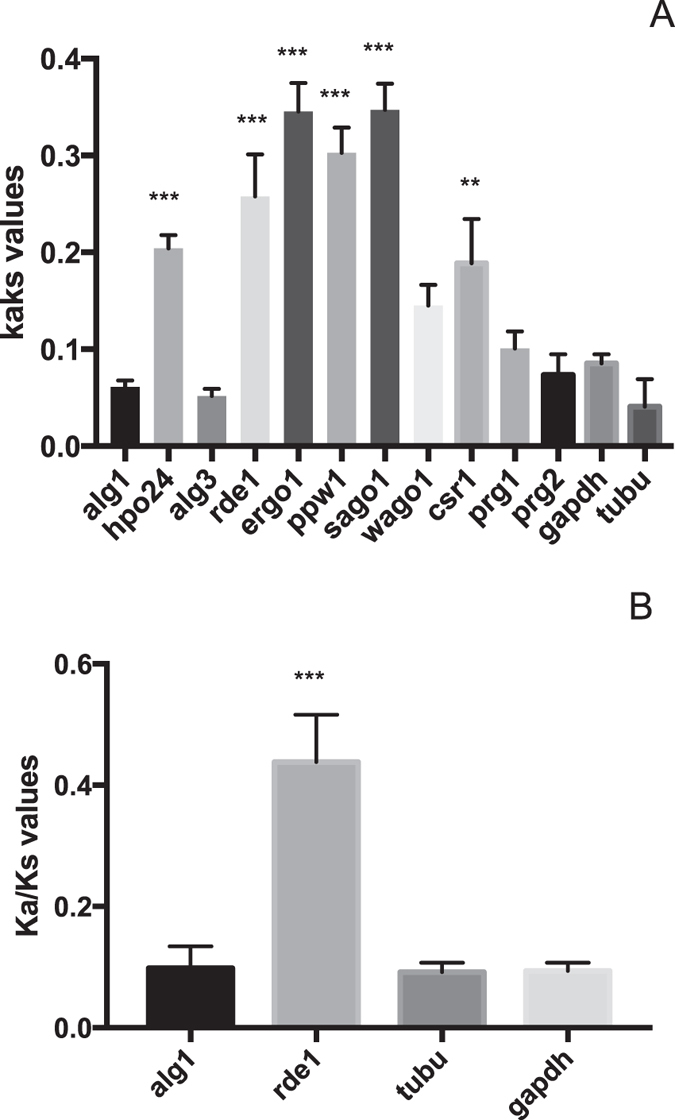
Figure 5A: Evolutionary rate of argonaute transcript sequences within the Caenorhabditis genus. Ka/Ks values are estimated by means of ML-tree based comparison and by using sequence information of C. elegans, C. briggsae, C. remanei and C. brenneri. Figure 5B: The evolutionary rate for Argonaute proteins in nematodes, determined by estimating the Ka/Ks values by means of ML-tree based comparison and by using sequence information of C. elegans, Ascaris suum, Brugia malayi and Necator americanus. Due to the enormous complexity and variation in the Argonaute AGO family present in nematodes, we could only find orthologs of the miRNA-class AGO alg1 and the siRNA-class AGO rde1 in all studied nematode species. The bars represent the mean +/− SEM of individual Ka/Ks values per gene. Statistics (MWU-test) support difference with the housekeeping genes glyceraldehyde 3-phosphate dehydrogenase (gapdh) and apha-tubulin1a (tubu); **p < 0.01, ***p < 0,0001.
