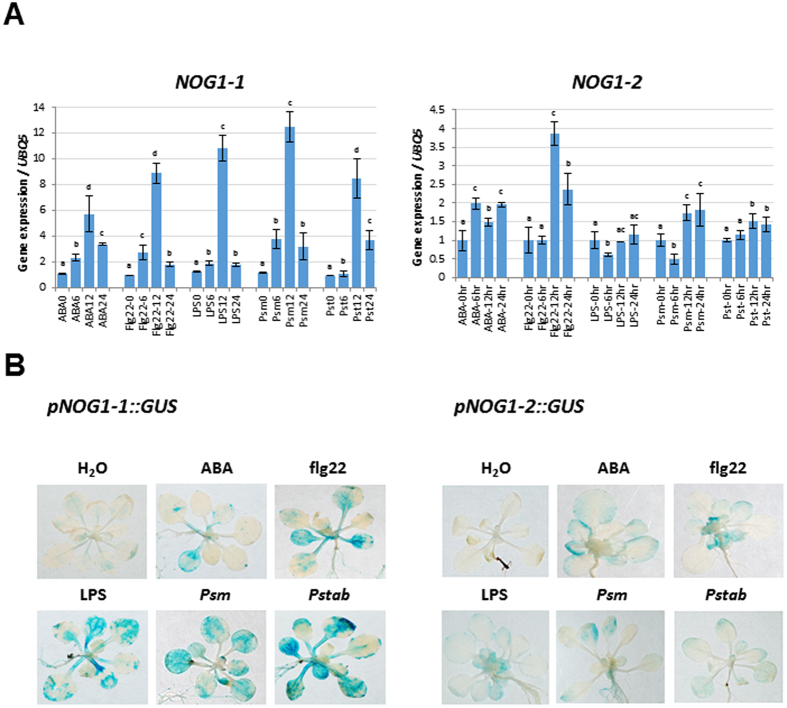
Figure 3.
NOG1-1 and NOG1-2 are induced by ABA, PAMPs, host and nonhost bacterial pathogens. (A) Arabidopsis wild-type (Col-0) plants were individually syringe-infiltrated with ABA (10 µM), Flg22 (20 µM), or LPS (100ng), or flood-inoculated with the pathogens P. syringae pv. maculicola (Psm) and P. syringae pv. tabaci (Pstab) at 1 × 104 CFU/ml. RNA was isolated from tissue samples harvested at 0 hr, 6 hr, 12 hr and 24 hr, and qRT-PCR was performed. Bars indicate relative gene expression in comparison with the housekeeping gene Ubiquitin (UBQ5) and in relation to 0 hr time that was considered as 1. Different letters above bars indicate a statistically significant difference within a treatment using two-way ANOVA (P < 0.01). Error bars represent the standard deviation of three biological replicates (three technical replicates for each biological replicate). (B) β-Glucuronidase (GUS) staining of pNOG1-1::GUS and pNOG1-2::GUS in response to ABA, PAMPs, bacterial pathogens. pNOG1-1::GUS (left panel) and pNOG1-2::GUS (right panel) expression was measured 12 hr after treatment with ABA, flg22, LPS, Psm and Pstab. Seedlings were flood-inoculated with both pathogens (1.4 × 106 CFU/ml), ABA (10 µM) and PMAPs (flg22: 20 µM, and LPS: 100ng). After 2 hr of GUS straining, plants were washed with sterile water and images were obtained.