Fig. 1.
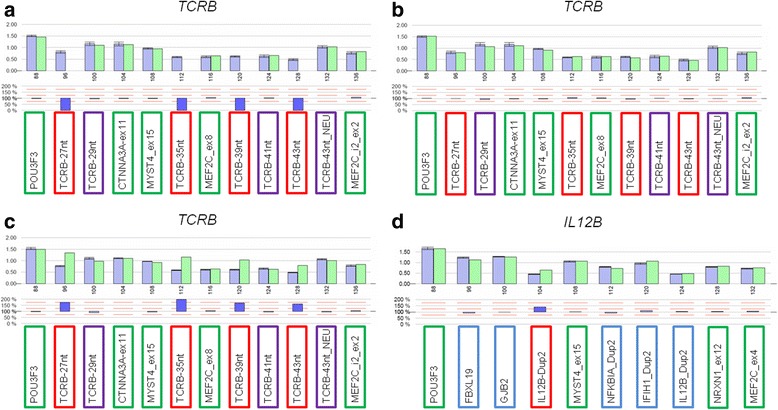
Investigation of copy number states by MLPA analysis. a-c: Copy number (CN) states at TRB, calibrated by carriers of one gene copy. The CNV region was targeted by four different MLPA probes (TCRB_27nt, TCRB_35nt, TCRB_39nt and TCRB_43nt; red frame), CN-stable regions encompassing the deleted region were covered by three additional MLPA probes (TCRB_29nt, TCRB_41nt and TCRB_45nt_NEU; purple frame). Five CN-stable control loci from other genomic regions (green frame) were included in the analysis as well (red and purple coloring corresponds to the respective probes in Fig. 2f). a Deletion in homozygous form, b Deletion in heterozygous form and c no deletion, i.e. two gene copies; d CN states at a CNV at the IL12B locus, calibrated by carriers of two gene copies; duplication targeted by one MLPA probe (IL12B_Dup2; red frame). Signals for four control loci located at CN-stable regions (green frame) and five CN variable regions (blue frame) were included in the analysis as well (data are from different PsA patients; one at IL12B [Dup1, not overlapping with Dup2], FBXL19, GJB2, NFKBIA and IFIH1, respectively)
