Fig. 2.
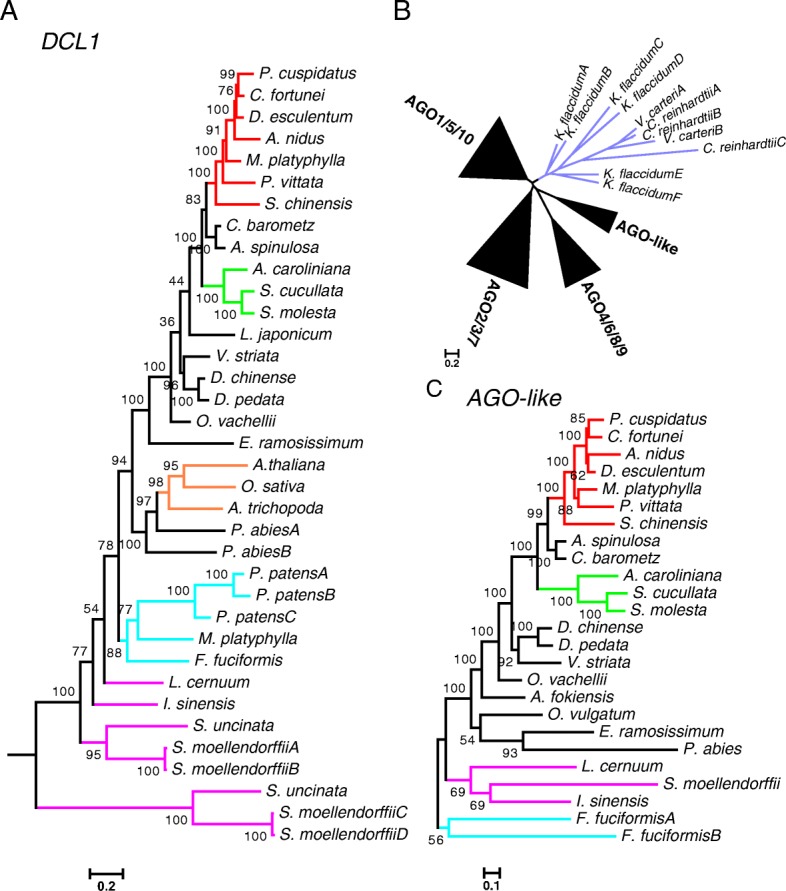
Phylogenetic trees of vital genes involved in small RNA pathways in land plants. a A maximun likelihood (ML) tree for DCL1 genes in representative land plants. The other subclades of DCL genes (such as DCL3 and DCL4) are shown in Additional file 2: Figure S1. Alignment length, 22,788 nt. b A schematic tree of AGO genes. The sequences from green algae (Chlamydomonas reinhardtii, Volvox carteri, and Klebsormidium flaccidum) are clustered. The AGO genes from land plants form four clades, three of which are named after the AGO genes from A. thaliana. c A ML tree for the AGO-like clade in b. Other AGO clades in land plants (such as AGO1/5/10) are shown in Additional file 2: Figure S2. Purple, green algae; light blue, bryophytes; magenta, lycophytes; red, Polypodiales (ferns); green, Salviniales (ferns); orange, angiosperms; black, other ferns and gymnosperms. The scale bar represents nucleotide substitution rates. Numbers beside nodes are bootstrap support values showing confidence (from 0 to 100). Alignment length, 3075 nt
