Fig. 2.
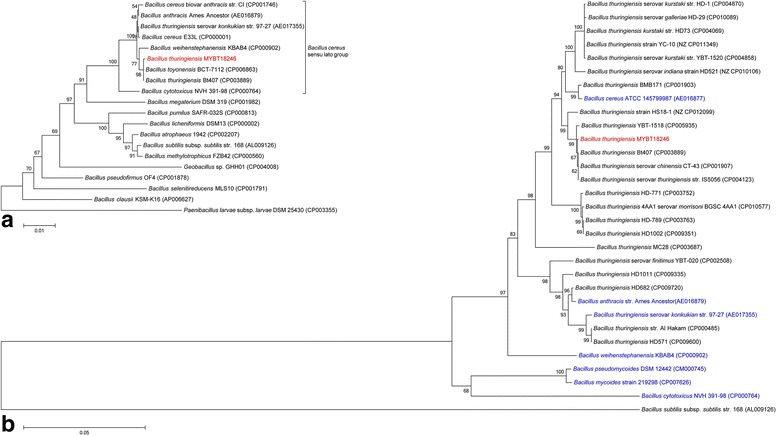
Phylogenetic tree highlighting the taxonomic relation of 10.1601/nm.5000 MYBT18246 (red) based on a) 16rDNA amplicon within the 10.1601/nm.4857 clade b) Multi-locus sequence typing within the 10.1601/nm.4885 sensu lato species group. GenBank accession numbers are given in parentheses. Comparison includes strains of the 10.1601/nm.4854 clade or Bcsl group members (blue). 10.1601/nm.5141 10.1601/strainfinder?urlappend=%3Fid%3DDSM+25430 or 10.1601/nm.4858 str. 168 has been used as outlier to root the tree. Sequences were aligned using ClustalW 1.6 [91, 92]. The phylogenetic tree was constructed by using the Neighbor-Joining method [50] and evolutionary distances were computed by the Maximum Composite Likelihood method [51] within MEGA7.0 [52]. Numbers at the nodes are bootstrap values calculated from 1000 replicates
